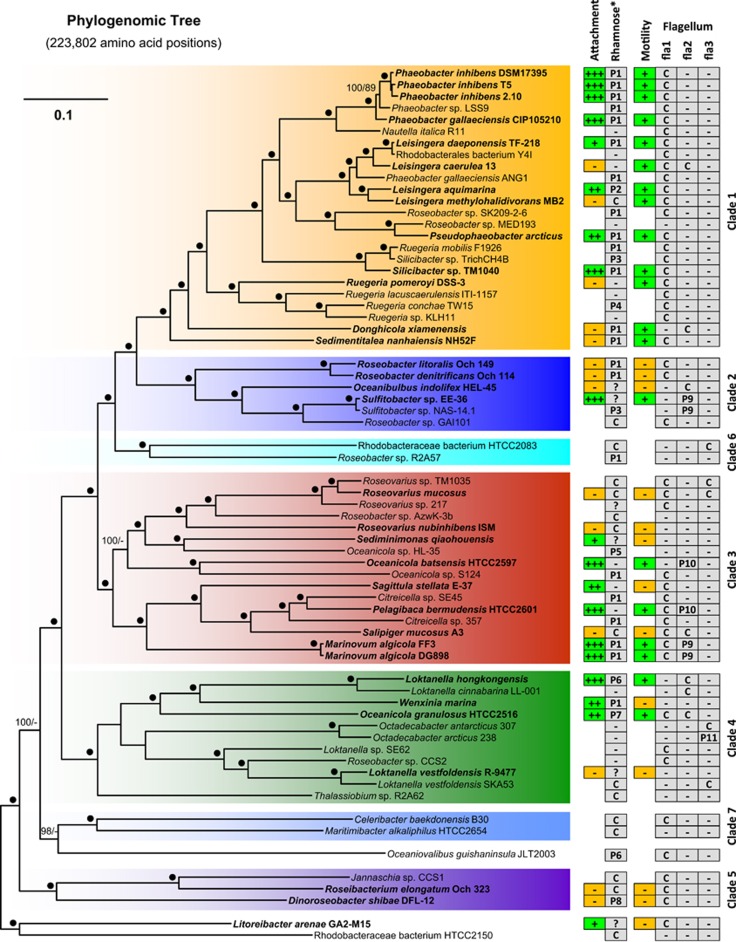
Figure 1.
Phylogenetic tree of the maximum-likelihood (ML) analysis of 67 sequenced Roseobacter genomes based on 223 802 amino-acid positions representing 643 genes of the core genome. The color code and numbering of the different clades refer to the phylogenetic tree of Newton et al. (2010). Strains that were used for attachment and motility assays are shown in bold. The branch lengths are proportional to the inferred number of substitutions per site. Statistical support for internal nodes of the ML tree was determined with rapid bootstrapping and the bootstopping criterion. For maximum parsimony (MP), 1000 replicates with 10 rounds of heuristic search per replicate were computed. Support values >60% are shown (ML, left; MP, right). ‘●', 100% bootstrap support ML and MP; ‘+++, ++, +', attachment and motility according to Table 1; ‘−', lacking ability of biofilm formation, motility (Table 1) or the absence of the rhamnose operon/flagellar gene cluster in the genome; ‘C', chromosomal localization; ‘P1–P11', plasmid localization (P1=RepA-I-type plasmid; P2=RepB-I, P3=RepABC-8, P4=DnaA-like II, P5=RepA-IV, P6=RepA*, P7=RepABC-3, P8=RepABC-1, P9=DnaA-like I, P10=RepABC-3, P11=RepB-III; Petersen et al., 2009; Petersen et al., 2011; Supplementary Table 1]); ‘?' localization unknown.