When inflammasomes trigger pyroptosis, the plasma membrane tears release-soluble cytosolic contents but retains organelles and intracellular bacteria. Neutrophils then efferocytose the pyroptotic cell debris and kill the entrapped bacteria.
Abstract
Inflammasomes activate caspase-1 in response to cytosolic contamination or perturbation. This inflammatory caspase triggers the opening of the GSDMD pore in the plasma membrane, resulting in lytic cell death called pyroptosis. We had previously assumed that pyroptosis releases intracellular bacteria to the extracellular space. Here, we find that viable bacteria instead remain trapped within the cellular debris of pyroptotic macrophages. This trapping appears to be an inevitable consequence of how osmotic lysis ruptures the plasma membrane, and may also apply to necroptosis and some forms of nonprogrammed necrosis. Although membrane tears release soluble cytosolic contents, they are small enough to retain organelles and bacteria. We call this structure the pore-induced intracellular trap (PIT), which is conceptually parallel to the neutrophil extracellular trap (NET). The PIT coordinates innate immune responses via complement and scavenger receptors to drive recruitment of and efferocytosis by neutrophils. Ultimately, this secondary phagocyte kills the bacteria. Hence, caspase-1–driven pore-induced cell death triggers a multifaceted defense against intracellular bacteria facilitated by trapping the pathogen within the cellular debris. Bona fide intracellular bacterial pathogens, such as Salmonella, must prevent or delay pyroptosis to avoid being trapped in the PIT and subsequently killed by neutrophils.
INTRODUCTION
Many pathogens target mammalian cells to establish intracellular replicative niches. In response, the host utilizes inflammasomes that detect cytosolic microbial ligands or perturbations. Three different bacterial proteins are detected in the host cytosol by the NLRC4 inflammasome: flagellin via the NAIP5/6 adaptor, T3SS rod (PrgJ in Salmonella SPI1) via NAIP2, and T3SS needle (PrgI in Salmonella SPI1) via NAIP1 (Kofoed and Vance, 2011; Zhao et al., 2011; Rayamajhi et al., 2013a; Yang et al., 2013). These three proteins are inefficiently translocated into the host cell cytosol by the SPI1 type III secretion system (T3SS) during infection with Salmonella enterica serovar Typhimurium (S. Typhimurium). The study of S. Typhimurium was instrumental in discovering these three NLRC4 agonists (Franchi et al., 2006; Miao et al., 2006, 2010b; Zhao et al., 2011; Rayamajhi et al., 2013a; Yang et al., 2013), but ironically, S. Typhimurium efficiently evades the NAIP/NLRC4 inflammasomes during systemic infection by repressing flagellin expression and switching from SPI1 to an NLRC4-evasive SPI2 T3SS (Miao and Rajan, 2011).
Inflammasomes oligomerize into multiprotein complexes, which serve as caspase-1 activating platforms. Caspase-1 mediates the secretion of the proinflammatory cytokines IL-1β and IL-18, and initiates a lytic and inflammatory form of programmed cell death (Zychlinsky et al., 1992), termed pyroptosis. After caspase-1 activation, pore formation in the plasma membrane causes water influx, cell swelling, and lysis. Pyroptosis is also induced by caspase-11, and is the likely mechanism by which caspase-11 defends against cytosolic-invasive Gram-negative bacteria (Aachoui et al., 2013, 2015). Removing the intracellular replicative niche is obviously beneficial to the host, which raises a key question: How does pyroptosis of infected macrophages ultimately lead to the clearance of the infection?
Mammalian cells can die via accidental cell death (necrosis) or via programmed cell death (e.g., pyroptosis, necroptosis, apoptosis, or NETosis). The term pyroptosis is coined from the Greek word pyro, meaning fire or fever, and ptosis, meaning falling (Cookson and Brennan, 2001), which reflects its inflammatory nature and highlights that it is distinct from other forms of programmed cell death. Necroptosis is induced by the activation of the RIP3 kinase, resulting in the opening of the MLKL pore, followed by swelling and lysis (Mocarski et al., 2015). Although distinct signaling events drive pyroptosis and necroptosis, they are similar in that they both trigger pore-induced lytic cell death programs. In contrast to pyroptosis and necroptosis, necrosis describes accidental, nonprogrammed cell death that results from physical cellular injury by external forces (Kroemer et al., 2009). Distinct from inflammatory forms of cell death, apoptosis is inherently noninflammatory and dependent on caspase-3 or -7. Apoptosis functions as part of normal development, maintenance of tissue homeostasis, and in protection against infection (Elmore, 2007). It is also characterized by the partitioning of cellular contents within membrane-bound apoptotic bodies. Finally, neutrophils can use yet another innate defense mechanism against extracellular pathogens termed neutrophil extracellular traps (NETs), used during a distinct form of programmed cell death called NETosis. Neutrophils release chromatin fibers covered in granules containing anti-microbial peptides and enzymes. NETs capture and kill extracellular bacteria (Brinkmann et al., 2004).
Regardless of the mechanism of cell death, the cellular debris must be removed to reestablish tissue homeostasis. The engulfment of apoptotic bodies and necrotic debris by professional phagocytes, termed efferocytosis, is essential in promoting tissue homeostasis (Poon et al., 2010). Efferocytosis of apoptotic bodies is driven by receptor binding of specific find-me signals, such as ATP, and eat-me signals, such as phosphatidyl serine, released or presented by the apoptotic body (Poon et al., 2010; Ravichandran, 2011). In contrast to clearance of apoptotic bodies, the mechanisms by which pyroptotic cellular remnants are removed, and how this participates in the resolution of infection, have not been studied.
Many bacteria, such as S. Typhimurium and Listeria monocytogenes, efficiently evade inflammasome detection in vivo (Jorgensen and Miao, 2015). Therefore, the best tool available to study pyroptosis and how it mediates bacterial clearance are engineered strains of S. Typhimurium where flagellin expression is experimentally controlled. We previously showed that such engineered strains are cleared via pyroptosis (WT S. Typhimurium evades this), involving the transfer of the bacterium from the primary macrophage undergoing pyroptosis to a secondary NADPH oxidase-expressing phagocyte (Miao et al., 2010a). However, the steps between pyroptosis and the killing of the bacterium by the secondary phagocyte remain unknown. Here, we study this process and find that the pyroptotic cellular debris has specific immunological functions that trap the previously intracellular bacteria, promoting uptake into neutrophils by efferocytosis.
RESULTS
Description of tools to probe pyroptosis in vitro and in vivo
The tools described herein are used to gain temporal control of inflammasome activation and pyroptosis to investigate the precise mechanisms of the downstream innate immune response. SPI1-induced S. Typhimurium (late log phase culture) express flagellin, PrgJ rod, and PrgI needle, which activate NLRC4 in vitro. In contrast, SPI1-repressed S. Typhimurium (overnight culture or intracellular growth) do not activate NLRC4 in vitro (Miao et al., 2010b). A second means to deliver flagellin to the cytosol is FlaTox, the anthrax lethal toxin with the catalytic domain replaced by flagellin, which serves as a vehicle to deliver flagellin to the cytosol independent of bacterial infection (von Moltke et al., 2012). We also use several other bacterial species, all of which are used under culture and infection conditions where they do not trigger caspase-1 activation (Tables S1 and S2 and Fig. 1 A).
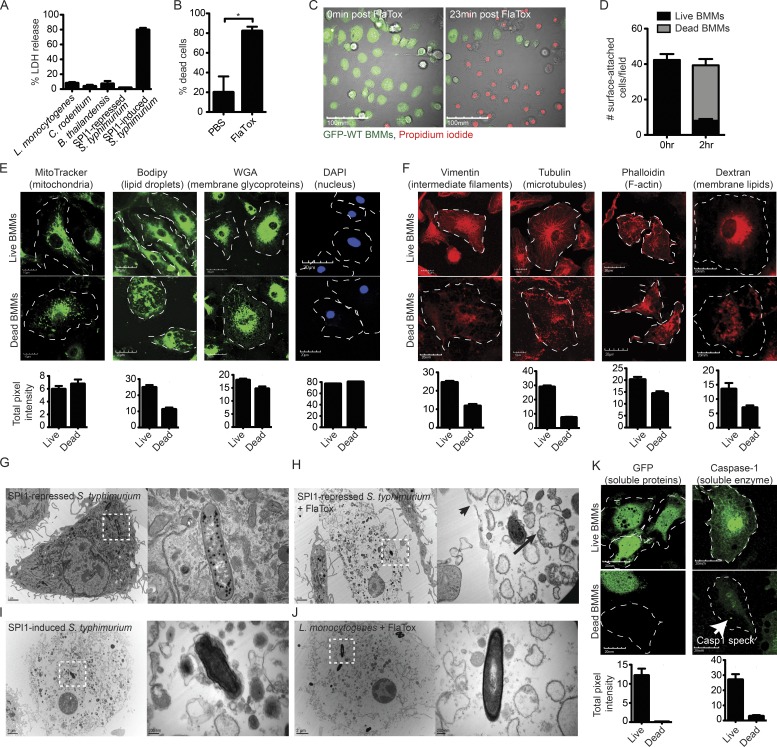
Figure 1.
The pyroptotic cell corpse consists of insoluble cellular contents. (A) WT BMMs were infected at an MOI of 20–25 for 2.5 h with L. monocytogenes, C. rodentium, B. thailandensis, or SPI1-repressed S. Typhimurium (under conditions where minimal inflammasome detection is observed) or with SPI-1–induced S. Typhimurium (which activates the NLRC4 inflammasome). Cell lysis was determined by LDH release. (B) WT BMMs were treated with PBS or 3 µg/ml FlaTox for 2 h. Percentage cell death was determined by counting based on morphology in DIC microscopy, which was equivalent to counting based on PI positivity (not depicted; C–F and K). BMMs were infected with SPI1-induced WT S. Typhimurium for 2.5 h (E) or treated with 3 µg/ml FlaTox for 2 h (C, D, F, and K), and then imaged by confocal microscopy. MitoTracker, Bodipy, WGA, dextran, and GFP labels were endogenous or added before treatment, and live cells were imaged. For vimentin, tubulin, phalloidin, caspase-1, and DAPI, cells were fixed and stained before imaging. Caspase-1–positive-speck in dead BMMs is indicated by a white arrow (K). Dead BMMs were identified by PI-staining (E and K) or DIC (F). Images are representative; quantitation of cellular markers was done in ImageJ by measuring the total intensity of ∼50 individual live and dead BMMs at time 0 and 2.5 h after infection or 2 h after FlaTox. (D) To determine the number of surface-attached cells before and after pyroptosis, the number of GFP (live) and PI (dead) cells from (C) were counted at time 0 and 2 h after FlaTox. (G–J) Electron micrographs of WT BMMs were infected with SPI1-repressed S. Typhimurium or L. monocytogenes followed by 3 µg/ml FlaTox treatment (G, H, and J) or infected with SPI1-induced S. Typhimurium (I). Frank rupture of the plasma membrane (arrowhead) and mitochondria with collapsed cristae (arrow). Dotted squares show individual bacteria, with greater magnification on right panel. Data are representative of two (A–F and K) and one (G–J) experiments. Error bars represent SE. P-values were determined by a two-tailed Student’s t test; *, P < 0.05.
During systemic infection in vivo, WT Salmonella represses flagellin and expresses the NLRC4-evasive SPI2 T3SS. This allows us to engineer the bacterium to induce expression of flagellin (FliCind), making the engineered strain a useful tool with which to interrogate inflammasome responses in vivo. To doubly ensure no chromosomal flagellin is expressed during experiments, we use a flgB mutant for in vivo studies with FliCind bacteria. Therefore, WT Salmonella avoids detection by inflammasomes, whereas flagellin-engineered Salmonella is cleared because it triggers the inflammasome response.
The pyroptotic cell corpse consists of insoluble cellular contents
What will phagocytes encounter once they arrive at the location where an infected macrophage has lysed via pyroptosis? During apoptosis, cellular contents are partitioned into membrane-bound apoptotic bodies. In contrast, lysis of a cell with strong chemical detergents or heat will fully disrupt all membranes in the cell. A detailed microscopic analysis revealed that lytic cell death via pyroptosis is not akin to either of these examples.
We visualized the process of pyroptosis by time-lapse confocal microscopy in WT BMMs. Pyroptosis was induced by infecting GFP-expressing BM-derived macrophages (BMMs) with SPI1-induced S. Typhimurium or by treatment with FlaTox, both of which activate NLRC4. The plasma membrane becomes permeable 20–30 min after infection, and ∼80% of all cells die by 2 h after infection (Fig. 1, A and B), as detected by propidium iodide (PI) entry (Fig. 1 C; Fink and Cookson, 2006). The cell corpse almost always remains attached to the glass or plastic substrate (Fig. 1 D). As the cell swells, membranous balloons rapidly expand and then rupture, resulting in the release of soluble cytosolic contents to the media, including proteins such as GFP (Video 1 and Fig. 1 C) and lactate dehydrogenase (LDH; Brennan and Cookson, 2000). It is important to note that this balloon-like swelling should not be called a bleb, as the term blebbing refers to a morphologically distinct process during apoptosis. In contrast to the rapid loss of soluble cytosolic proteins, the insoluble components of the cell remain associated with each other. Many organelles are retained within this pyroptotic cell corpse, as demonstrated by live cell or post-fixation labeling of organelles, including vacuolar membranes (Dextran and wheat germ agglutinin [WGA]), mitochondria (MitoTracker), lipid droplets (Bodipy), the nucleus (DAPI), the actin cytoskeleton (phalloidin), microtubules (tubulin), and intermediate filaments (vimentin; Fig. 1, E–F). Organelle stains dim to a certain extent after pyroptosis, which may be a result of the cell moving lower in the focal plane after pyroptosis or a loss of a fraction of this compartment. The nucleus, however, always remains associated with the pyroptotic cell corpse, and DNA content remains unchanged (Fig. 1 E).
Ultrastructural analysis by electron microscopy (EM) of WT BMMs induced to undergo pyroptosis further demonstrated the association of insoluble material with the cell corpse. Similar to confocal microscopy, representative EM images showed an intact cell corpse with a condensed nucleus and associated bacteria (Fig. 1, G–J). Organelle debris appears swollen with a complete loss of structure, as indicated by confocal microscopy. Organelle structure is damaged, even though the bulk of organelles remain associated with the pyroptotic cell corpse. For example, only remnants of the mitochondrial crista (Fig. 1 H, long black arrow) can be discerned within the still membrane-bound organelle (Fig. 1, G–J), and the actin, intermediate filament, and microtubule cytoskeleton collapse (Fig. 1 F). Nevertheless, a significant fraction of cytoskeletal markers remains after pyroptosis, in contrast to soluble proteins such as GFP and caspase-1, which are completely lost with the exception of caspase-1 bound to the polymerized caspase-1-speck (Fig. 1 K, white arrow).
Pyroptotic cell corpses induced by either FlaTox or SPI1-induced S. Typhimurium look structurally identical (Fig. 1, H and I). Although a relatively large tear ranging from ∼0.1 to 0.25 µm can be found, the plasma membrane remains largely intact, retaining large remnants of insoluble membrane-bound organelles.
The pyroptotic cell corpse traps live bacteria
After pyroptosis, we frequently observed that bacteria remained within the cell corpse (Fig. 2 A and Video 2), suggesting that the largely intact plasma membrane entraps intracellular bacteria. Images of cell corpses in the z plane confirmed that the bacteria reside within the cell corpse and are not simply attached to the extracellular surface (Fig. 2 B). This was independent of whether BMMs had been infected for 2 or 24 h with SPI1-repressed S. Typhimurium when treated with FlaTox (Fig. 2 C). Although some bacteria appear to retain limited mobility within the pyroptotic cell debris (Videos 2), the majority of bacteria are immobilized. The cytoskeleton is not required for this trapping, as disrupting microtubules (nocodazole) or the actin cytoskeleton (cytochalasin D) does not prevent trapping (Fig. 2 A).
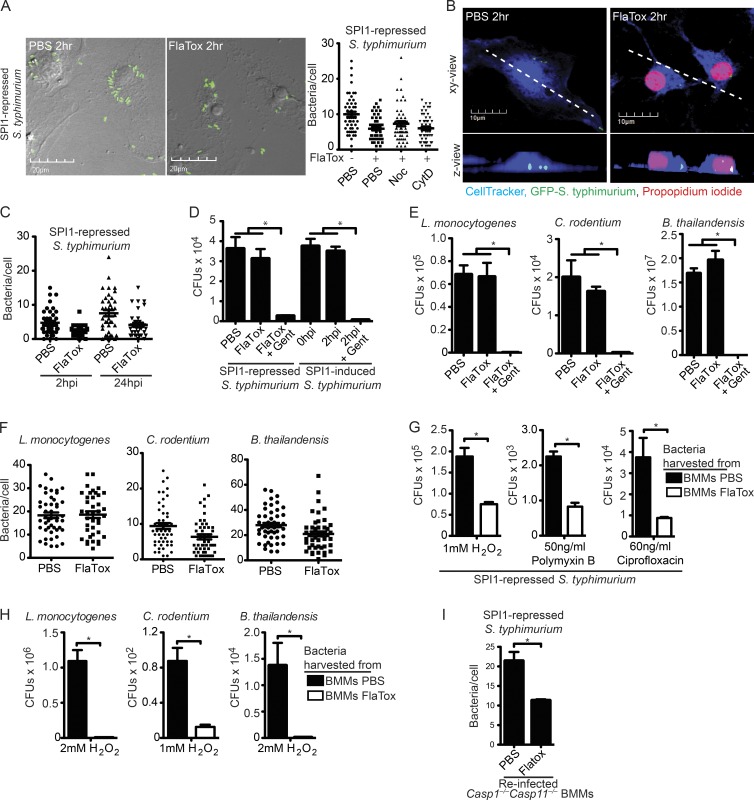
Figure 2.
The pyroptotic cell corpse traps live bacteria. (A–I) WT BMMs were infected with the indicated inflammasome-nonactivating bacteria, followed by FlaTox as in Fig. 1. Alternately, they were infected with inflammasome-activating SPI-1–induced S. Typhimurium as in Fig. 1. (A, C, F, and I) To examine bacterial association with pyroptotic cell debris, the number of bacteria/cell within live (PBS) and dead (FlaTox) BMMs was quantified by counting cell-associated bacteria in ∼100 cells from six fields in a single plane of images acquired by confocal microscopy. (D, E, G, and H) To determine bacterial viability, CFUs were enumerated by plating. (A) Nocodazole or cytochalasin D were added during FlaTox treatment. (B) BMMs were labeled with CellTracker Blue. Z-stacks acquired by confocal-microscopy were used for 3D reconstruction of live and dead BMMs and to generate the z view. (C) Longer resident time within the BMM does not alter the bacterial viability after pyroptosis. (D–E) Gentamicin was added during FlaTox treatment. (G and H) Infected, treated cells were then lysed and cell lysates were added to LB media and incubated in the presence of 1 or 2 mM H2O2, 50 ng/ml Polymyxin B, or 60 ng/ml ciprofloxacin for 2 h, and CFUs were enumerated by plating. (I) Cells were collected by scraping, and then lysed with a 30-gauge needle. Casp1−/−Casp11−/− BMMs were infected with cellular lysates, washed, treated with gentamicin for 2 h, fixed, and imaged. The number of Casp1−/−Casp11−/− BMMs with GFP-containing bacteria was determined for ∼100 individual cells from 10 fields. All data are representative of three individual experiments. All error bars represent SE. P-values were determined by a two-tailed Student’s t test; *, P < 0.05.
To determine if bacteria trapped in pyroptotic cell corpses are viable, we infected BMMs with SPI1-repressed S. Typhimurium to establish an intracellular population. Subsequent induction of pyroptosis with FlaTox did not alter bacterial viability (Fig. 2 D). Survival after pyroptosis was independent of the method of flagellin delivery to the cytosol, as S. Typhimurium remained viable after FlaTox- and SPI1-induced pyroptosis (Fig. 2 D). However, the pyroptotic cell debris–entrapped bacteria became exposed to gentamicin, a membrane impermeant antibiotic (Fig. 2 D). These observations extend to all intracellular bacteria tested, such as Listeria monocytogenes, Citrobacter rodentium, and Burkholderia thailandensis (Fig. 2 E), the vast majority of which remained associated with the pyroptotic cell corpse (Fig. 2 F).
Therefore, we propose that the ruptured but nevertheless largely intact plasma membrane (Fig. 1, G–J) functionally traps bacteria, holding them within the corpse of the cell after pyroptosis.
Pyroptosis damages bacteria
EM images of S. Typhimurium associated with pyroptotic cell corpses revealed a denser and slightly ruffled appearance of the bacterial outer membrane compared with bacteria within live BMMs (Fig. 1, G–I). This suggested that the bacterial outer membrane might be structurally damaged after pyroptosis, although not severely enough to kill the bacteria (Fig. 2, D and E). We therefore hypothesized that pyroptosis-induced damage could cause the bacteria to be more susceptible to other stressors compared with bacteria in intact nonpyroptotic macrophages.
We harvested bacteria from intact BMMs and pyroptotic cell corpses, and examined susceptibility to ROS (H2O2), an antimicrobial peptide (polymyxin B), and an antibiotic (ciprofloxacin). S. Typhimurium (SPI1-repressed) associated with FlaTox-induced cell corpses were significantly more susceptible to H2O2, polymyxin B, and ciprofloxacin compared with bacteria from intact BMMs (Fig. 2 G). Increased susceptibility to H2O2 was observed for all other intracellular bacteria tested, such as L. monocytogenes, C. rodentium, and B. thailandensis (Fig. 2 H). Interestingly, the cell wall of L. monocytogenes within a pyroptotic cell corpse was not visibly altered by EM (Fig. 1 J), perhaps because of the thicker peptidoglycan layer in the Gram-positive cell wall.
Finally, we investigated whether pyroptosis-induced damage rendered the bacteria less capable of reinfecting a macrophage. Indeed, fewer cell corpse–associated S. Typhimurium were recovered from a brief secondary infection of Casp1−/−Casp11−/− BMMs compared with bacteria from intact BMMs (Fig. 2 I), suggesting that pyroptosis decreases the infectivity of the pathogen. Thus, pyroptosis fails to kill bacteria, but may render them more susceptible to subsequent attack.
Necrosis and necroptosis result in similar cell corpses
To determine if the pyroptotic cell corpse is structurally similar to the cell corpse generated by other forms of lytic cell death, WT BMMs were treated with FlaTox, the mild detergent saponin, or TNF/BV6/ZVAD to induce pyroptosis, necrosis, and necroptosis, respectively. Compared with other stronger detergents that completely solubilize lipid membranes, treatment with a low concentration of saponin generates pores in the plasma membranes, leading to cell swelling and subsequent lysis. Similar to the pyroptotic cell corpse, both the TNF/BV6/ZVAD and saponin pore-induced cell corpse retained insoluble cellular contents, such as vacuolar compartments and the nucleus, but completely lost the soluble compartment (GFP; Fig. 3, A–C). In contrast, apoptotic bodies retained the soluble compartment (GFP) in addition to the vacuolar compartment and nucleus (Fig. 3 D). As with pyroptosis, bacteria remained viable and associated with the dead cell corpse after the induction of necrosis, necroptosis, and apoptosis (Fig. 3, E–G).
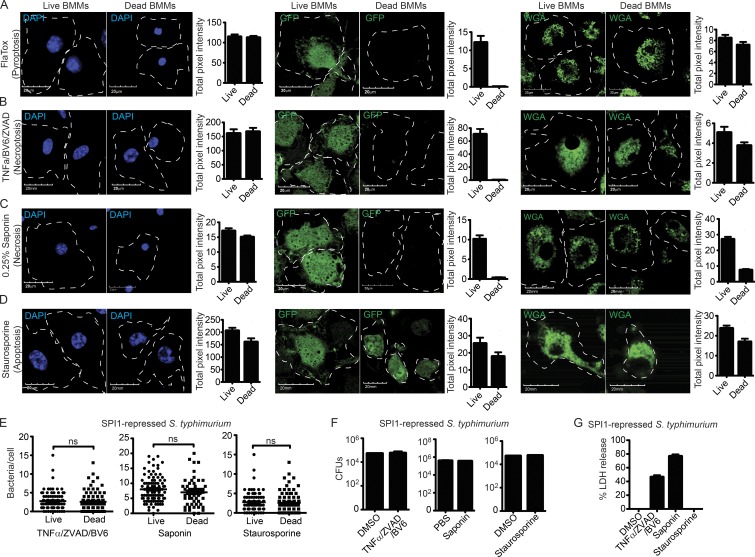
Figure 3.
Necrosis and necroptosis result in similar cell corpses. (A–G) WT BMMs were treated with (A) 3 µg/ml FlaTox, (B) 50 ng/ml TNF, 10 µM ZVAD, and 5 µM BV6, (C) 0.05% Saponin, or (D) 1 µM staurosporine for 2 h. (A–D) WGA-labeled, GFP-expressing, or fixed and DAPI-stained cells were imaged by confocal microscopy at 0 and 2 h after treatment. Quantitation of cellular markers was done in ImageJ by measuring the total intensity of ∼50 individual live and dead BMMs. (E–G) BMMs were infected with SPI-1 repressed GFP-expressing S. Typhimurium for 2.5 h before indicated treatment. (E) The number of bacteria/cell or viable CFU were determined as in Fig. 2. (G) Cell lysis by LDH release. Data are representative of two (A–D) and three (E–G) individual experiments. Error bars represent SE. P-values were determined by a two-tailed Student’s t test.
The nature of the cellular contents retained or lost is thus similar between all pore-induced cell death mechanisms (pyroptosis, necroptosis, and necrosis), but distinct from apoptosis (where GFP is retained). This pore-induced cellular debris has a plasma membrane that remains largely intact, trapping intracellular bacteria. Because this structure has specific immunological functions, further described below, we termed it the pore-induced intracellular trap (PIT).
Efferocytosis of pyroptotic cell corpses and entrapped bacteria in vitro
In addition to trapping the bacteria, the PIT may also present eat-me signals that promote efferocytosis of the cell corpse and associated bacterium by a secondary phagocyte. Indeed, we frequently observed uptake of the PIT by neighboring cells in video microscopy (Fig. 4 A and Video 3). When this occurs, sometimes the entire PIT and entrapped bacteria are consumed (Fig. 4 A and Video 4). Other times, large fragments with entrapped bacteria are physically ripped off of the main pyroptotic cell corpse (Video 3), and different fragments of the PIT can be efferocytosed by multiple neighboring macrophages (Video 5).
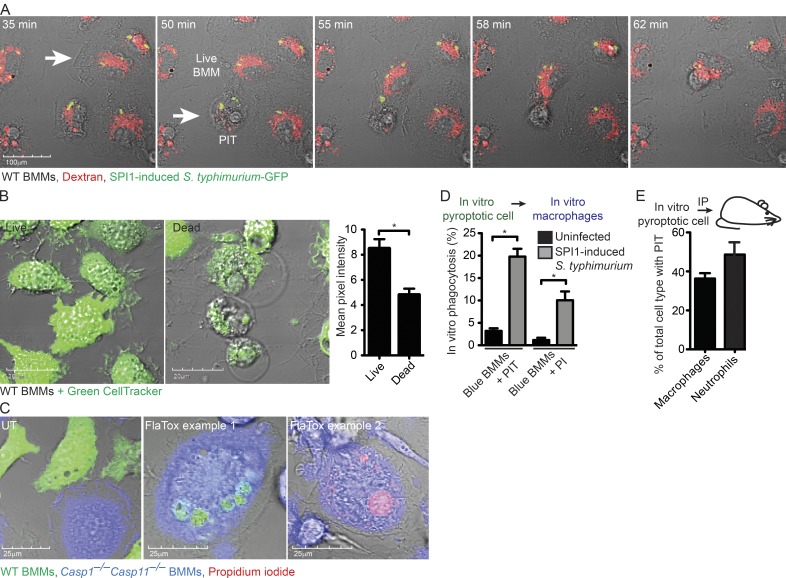
Figure 4.
Efferocytosis of PIT and associated bacteria in vitro. (A) Dextran–Alexa Fluor 555–labeled WT BMMs were infected with SPI1-induced mCherry-expressing S. Typhimurium as in Video 4 and imaged by live cell microscopy. Images are stills from Video 4 at indicated time points after infection. White arrow indicated a live BMM that efferocytoses a pyroptotic BMM denoted by the arrowhead. (B) WT BMMs were treated with PBS or 3 µg/ml FlaTox for 2 h, labeled with Green CellTracker and quantitated in ImageJ by measuring the total intensity of ∼50 individual live and dead BMMs. (C and D) WT BMMs (Green CellTracker) and Casp1−/−Casp11−/− BMMs (Blue CellTracker) were co-cultured and treated with 3 µg/ml FlaTox in PI-containing media while imaged by live cell confocal microscopy. The in vitro percentage of phagocytosis was determined by quantifying the percentage of Casp1−/−Casp11−/− BMMs (Blue) that contain CellTracker Green–BMM debris or PI-positive nuclei. (E) CellTracker Orange–labeled and FlaTox-treated WT BMMs were injected i.p into Casp1−/−Casp11−/− mice (n = 5). The percentage of live peritoneal macrophages (CD45+ CD11b+ Ly6G− F4/80+) and neutrophils (CD45+ CD11b+ Ly6G+ F4/80−) with CellTracker Orange-positive debris was determined by staining and analyzing the peritoneal wash by flow cytometry. All data are representative of three individual experiments. Error bars represent SE. P-values were determined by a two-tailed Student’s t test; *, P < 0.05.
To distinguish the pyroptotic cell corpse from the secondary phagocyte and to quantitate efferocytosis in vitro, we co-cultured pyroptosis-competent and -resistant BMMs and induced pyroptosis with FlaTox or SPI1-induced S. Typhimurium. Pyroptosis-competent WT BMMs were labeled with CellTracker Green before the induction of pyroptosis, and retained about half of the CellTracker signal after pyroptosis (Fig. 4 B). Pyroptosis-resistant Casp1−/−Casp11−/− BMMs were labeled with CellTracker Blue. PI was included to label the nucleus of the pyroptotic cell corpse. We then identified blue Casp1−/−Casp11−/− BMMs containing green-labeled PIT or PI-positive nuclei (Fig. 4 C). 2 h after inducing pyroptosis, 10 and 20% of Casp1−/−Casp11−/− BMMs had efferocytosed the green-labeled PIT or the PI-positive nuclei, respectively (Fig. 4 D). To determine the fate of pyroptotic cell corpses in vivo, we induced pyroptosis in CellTracker-labeled WT BMMs with FlaTox in vitro, and injected the pyroptotic cell corpse i.p. into mice. Flow cytometry analysis showed that 38% of macrophages and 49% of neutrophils had efferocytosed PITs (Fig. 4 E), indicating that phagocytes are capable of efferocytosing PITs, and that in vivo neutrophils are important in PIT efferocytosis.
Visualization of neutrophils efferocytosis of PIT in vivo
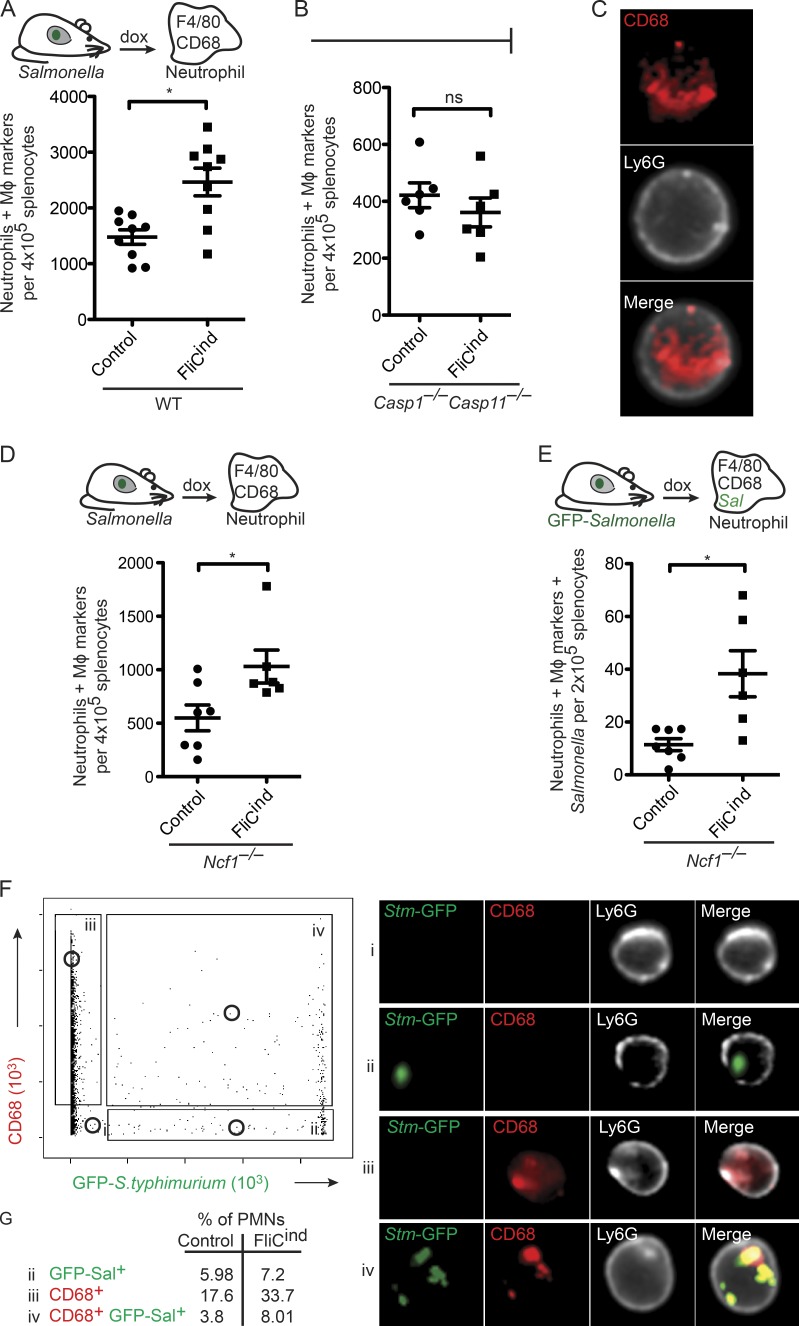
Although neighboring macrophages can efferocytose PITs in vitro, we have previously suggested that the physiologically relevant secondary phagocyte in vivo is the neutrophil (Miao et al., 2010a). To demonstrate that PITs are efferocytosed in vivo, we developed a flow cytometry-based assay to identify PIT-associated markers and/or GFP-tagged S. Typhimurium present within neutrophils (Fig. S1 A). To synchronize the induction of pyroptosis in vivo, we used our GFP-expressing S. Typhimurium strain where flagellin is expressed under a doxycycline-inducible promoter (FliCind). WT mice infected with either control or FliCind S. Typhimurium were treated with doxycycline for 3.5 h to induce pyroptosis in infected mice. Then, splenocytes were stained with neutrophil markers, fixed and permeabilized, followed by staining with macrophage markers F4/80 and/or CD68. Gating on these markers will identify neutrophils that contain PITs. Indeed, in comparison to mice infected with the control strain, there is a significant increase in the number of neutrophils with intracellular macrophage markers in mice infected with FliCind S. Typhimurium, indicating that pyroptosis induces efferocytosis of PITs (Fig. 5 A). As expected, this is not seen in Casp1−/−Casp11−/− mice, which cannot detect the induced flagellin (Fig. 5 B).
Figure 5.
Efferocytosis of PIT and entrapped bacteria in vivo. Spleens from mice infected i.p. with control or FliCind GFP–S. Typhimurium for 24 h and treated with doxycycline for 3.5 h, were harvested, prepared for flow cytometry, and stained. Neutrophils with intracellular macrophage (MΦ) markers (CD45+ CD11b+ Ly6G+high CD68+ F4/80+; A, B, and D), and neutrophils with intracellular MΦ markers and GFP–S. Typhimurium (CD45+ CD11b+ Ly6G+high CD68+ F4/80+ GFP+; E) were identified by flow cytometry on an LSR II. (C, F, and G) Neutrophils were isolated by purification of Ly6G+ cells from spleen, stained, and analyzed by Amnis ImageStream. Experimental data from WT (six mice per group) and Ncf1−/− mice (seven per control group and six per FliCind group) was pooled from two individual experiments. Data from six Casp1−/−Casp11−/− mice is representative of two individual experiments. The numbers of mice in all groups are noted in figure. Five Casp1−/−Casp11−/− mice were used per group and six animals per group were used for all other experiments. Control and FliCind GFP–S. Typhimurium strains are flagellin (flgB) mutants. Gating was performed using FlowJo (A, B, D, and E) or IDEAS (C, F, and G). P-values were determined by a two-tailed Student’s t test; *, P < 0.05.
To further demonstrate that CD68+ neutrophils identified on the LSR II flow cytometer (Fig. 5 A) represent bona fide phagocytosis of PITs by neutrophils, and to exclude the possibility of nonspecific adhesion of macrophage cellular debris to the exterior of the neutrophil, we used Amnis ImageStream. This technology combines flow cytometry with fluorescent imaging, taking a fluorescent picture of each cell that passes through the flow chamber. Mice were infected with vector control or FliCind and isolated neutrophils were analyzed on the ImageStream. Among neutrophils that stained for the CD68 macrophage marker, 93.4% had the CD68 staining in the interior of the cell (Fig. 5 C), consistent with efferocytosis of the PIT and arguing against nonspecific adhesion of macrophage debris to the exterior of the cell (Fig. 5 C).
Our in vitro data suggests that the secondary phagocyte will simultaneously efferocytose the bacteria entrapped within the PIT. We therefore hypothesized that neutrophils containing intracellular macrophage markers should also contain bacteria. However, once efferocytosed by neutrophils, the bacteria are rapidly killed, and thus difficult to experimentally detect. We therefore used NADPH oxidase-deficient mice (Ncf1−/−; also known as p47phox−/−), whose inability to kill FliCind bacteria results in accumulation within neutrophils after pyroptosis (Miao et al., 2010a). Using this strategy, we were able to again detect neutrophils that had phagocytosed macrophage markers after doxycycline injection using the LSR II flow cytometer (Fig. 5 D), and further, a subset of these neutrophils contained both macrophage markers and bacteria (Fig. 5 E). This suggests that the entrapped bacteria were cophagocytosed with the PIT.
We also visualized these cells on the ImageStream. Neutrophils were positive for the CD68 macrophage marker (gate iii), GFP-expressing Salmonella (gate ii), both (gate iv), or neither (gate i; Fig. 5 F). Neutrophils containing CD68 and GFP were 2.1 times more prevalent in FliCind-infected mice compared with vector controls (Fig. 5 G), consistent with the data obtained with the LSR II flow cytometer (Fig. 5 E). Although neutrophils containing CD68 and GFP could also be visualized with WT S. Typhimurium, these were much less frequent, which is consistent with the model that WT S. Typhimurium minimizes or delays macrophage cell death. More importantly, bacteria colocalized with the macrophage marker within neutrophils, and both were largely contained within the boundaries of the neutrophil (Fig. 5 F). Consistent with our observation that phagocytes can rip off large fragments of the PIT, which may or may not contain bacteria, and that macrophages are larger than neutrophils, we also observed a significant number of neutrophils positive for PIT markers but lacking bacteria (gate iii).
We therefore conclude that intracellular bacteria are trapped within the PIT and that neutrophils efferocytose the PIT (and associated bacteria) in vivo.
Role of neutrophils in killing PIT-associated bacteria
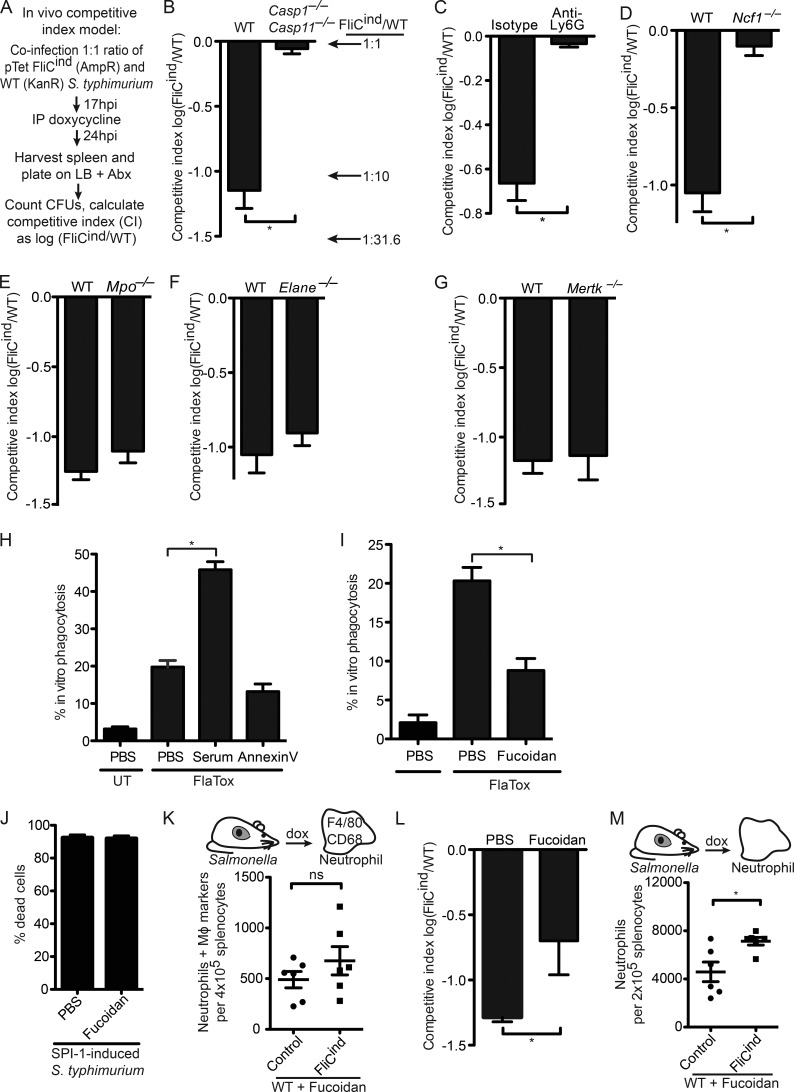
We next set out to study the effector mechanisms engaged by neutrophils to clear PITs, and their entrapped bacteria during in vivo infection by efferocytosis. To quantitate bacterial clearance after pyroptosis, we used the competitive index assay, where mice were co-infected with a 1:1 ratio of two S. Typhimurium strains: FliCind (ampR) and WT (kanR). This low dose infection was established for 17 h, during which time both strains evaded inflammasome detection and were phenotypically identical. Next, mice were injected with doxycycline to induce flagellin expression in FliCind; bacterial burdens were determined 7 h later (Fig. 6 A). The competitive index (CI) was expressed as the log of FliCind/WT CFUs, such that this 10-fold reduction in FliCind CFUs was expressed as a CI of −1.0. Upon splenic harvest, we recovered ∼10-fold less FliCind than WT CFUs from the spleen of WT mice, whereas Casp1−/−Casp11−/− mice failed to clear FliCind (Fig. 6 B), consistent with our previous results (Miao et al., 2010a). This defect was also observed in liver and draining lymph nodes (unpublished data). Because we observed efferocytosis of PITs in vivo by neutrophils, we further substantiated the role of neutrophils in this clearance by treating mice with anti-Ly6G or isotype control antibodies. Neutrophil depletion abrogated the clearance of FliCind S. Typhimurium (Fig. 6 C and Fig. S1 B). This neutrophil-mediated clearance was entirely dependent on ROS-mediated killing of bacteria, as Ncf1−/− mice failed to clear FliCind (Fig. 6 D), consistent with our prior data (Miao et al., 2010a).
Figure 6.
SRs mediate efferocytosis of PITs in vivo. (A–G and L) Mice were infected with 1:1 ratio of FliCind AmpR and WT KanR S. Typhimurium i.p. for 17 h, and treated with doxycycline. Spleens were harvested 7 h later. (C) Mice were injected with isotype or anti-Ly6G antibodies and infected 12 h after antibody treatment. (L) Mice were treated with Fucoidan at the same time as doxycycline. (H and I) WT BMMs (CellTracker Green) and Casp-1−/−Casp-11−/− BMMs (CellTracker Blue) were co-cultured, imaged and analyzed as in Fig. 4 C, and treated with serum (H), Annexin V (H), or fucoidan (I). (J) WT BMMs were infected with SPI1-induced S. Typhimurium as in Fig. 1 and treated with PBS or fucoidan for 2 h. Percentage dead cells was determined as in Fig. 1 B. (K and M) Spleens from mice (six animals per group) infected i.p with control or FliCind GFP–S. Typhimurium for 24 h and treated with doxycycline and fucoidan for 3.5 h, were harvested, prepared for flow cytometry and stained. Neutrophils with intracellular macrophage markers (CD45+ CD11b+ Ly6G+high CD68+ F4/80+) were identified by flow cytometry. All gating was performed using FlowJo. Data are representative of three individual experiments (A–J and L) or was pooled from two individual experiments (K and M). All error bars represent SE. P-values were determined by a two-tailed Student’s t test; *, P < 0.05.
Our model proposes that PITs defend against intracellular bacteria, and thus we hypothesized that NETs would not be involved as they target extracellular bacteria. Indeed, myeloperoxidase- and elastase-deficient mice successfully cleared FliCind (Fig. 6, E and F), despite their inability to form NETs (Metzler et al., 2011; Kolaczkowska et al., 2015). This was consistent with efferocytosis of PITs by neutrophils, which kill the internalized bacteria.
Scavenger receptors (SRs) recognize PITs and mediate efferocytosis in vivo
We next wanted to identify the neutrophil receptors and ligands that mediate efferocytosis. The apoptotic body presents eat-me ligands, including phosphatidyl serine (PS), which is recognized by MERTK and SRs (Peter et al., 2010; Poon et al., 2010). However, Mertk−/− mice cleared FliCind S. Typhimurium similar to WT mice in vivo (Fig. 6 G) and blocking PS with Annexin V protein did not prevent efferocytosis of PITs in vitro (Fig. 6 H). This suggests that the eat-me ligands presented on PITs are different from those that drive apoptotic cell efferocytosis.
SRs are a heterogeneous class of phagocytic receptors divided into several subclasses based on structural and functional properties (Murphy et al., 2005). The five members of the class A SRs (SR-A) can function as phagocytic receptors for both necrotic debris and apoptotic bodies (Canton et al., 2013). Fucoidan is a complex sulfated polysaccharide that has been used as a general inhibitor of the five SR-A family receptors. Fucoidan decreased efferocytosis of pyroptotic macrophages in vitro (Fig. 6 I); as a control, it did not prevent pyroptosis in vitro (Fig. 6 J). We next used the flow cytometry–based assay to assess if SRs mediate efferocytosis of PITs in vivo. Indeed, fucoidan reduced the number of neutrophils that efferocytosed PITs in vivo (Fig. 6 K), suggesting that neutrophils use SR-As to efferocytose PITs. Moreover, even though we observed in our CI assay that fucoidan reduced clearance of FliCind bacteria, there was residual clearance by other mechanisms (Fig. 6 L). Fucoidan also inhibits P- and l-selectin (Carvalho et al., 2014); however, we observed that pyroptosis still induced neutrophil influx to the spleen in the presence of fucoidan (Fig. 6 M). In summary, these results indicate that other phagocytic receptors likely act redundantly with SR-As.
Complement facilitates efferocytosis of PITs in vivo
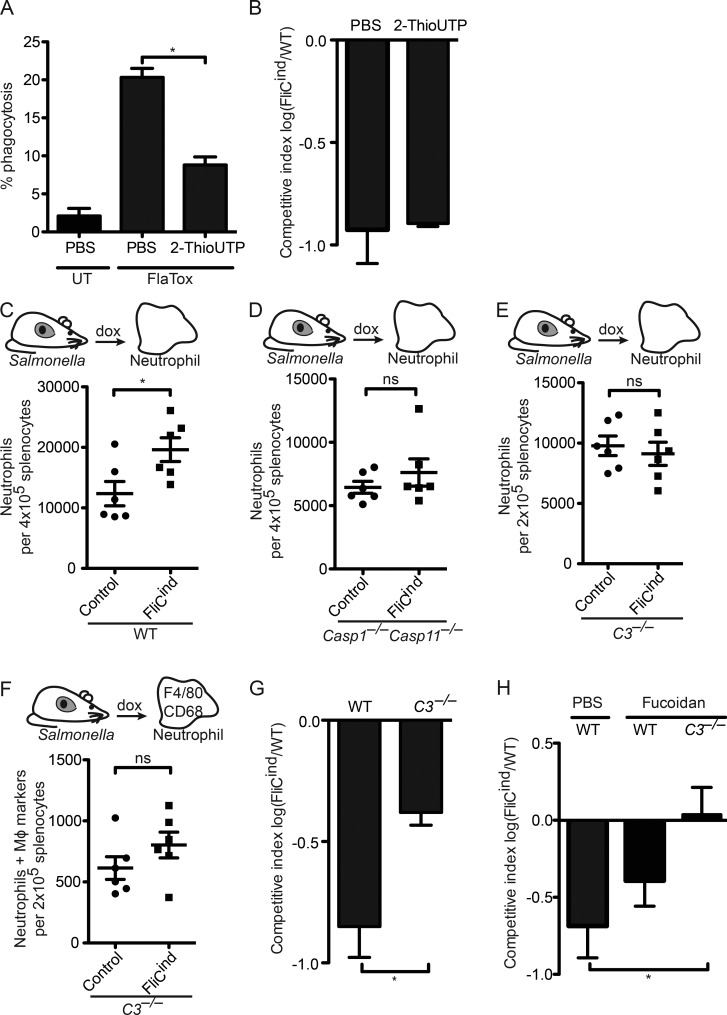
After extravasation of immune cells from the vasculature, the neutrophil must chemotax toward the site of the PIT, yet the signals that mediate neutrophil recruitment to the PIT location are unknown. ATP is the chemotactic find-me signal released during apoptosis, mediating chemotaxis by binding to the P2Y2 purinergic receptor on secondary phagocytes. In vitro, there is a small, but significant, decrease in the percentage of phagocytosis by Casp1−/−Casp11−/− BMMs treated with the P2Y2 antagonist 2-ThioUTP (Fig. 7 A). However, FliCind S. Typhimurium was cleared in WT mice treated with 2-ThioUTP (Fig. 7 B), suggesting that ATP is not essential in clearance of PITs and associated bacteria after pyroptosis.
Figure 7.
Complement facilitates efferocytosis of PITs in vivo. (A) WT BMMs (CellTracker Green) and Casp-1−/−Casp-11−/− BMMs (CellTracker Blue) were co-cultured, imaged, and analyzed as in Fig. 4 C, and treated with 2-ThioUTP. (B, G, and H) Mice (five animals per group) were infected with 1:1 ratio of FliCind AmpR and WT KanR S. Typhimurium IP for 17 h before doxycycline, fucoidan, and 2-ThioUTP injection; organs were harvested 7 h later. (C–F) Spleens from mice (six animals per group) infected IP with control or FliCind GFP–S. Typhimurium for 24 h and treated with doxycycline and fucoidan for 3.5 h were harvested and prepared for flow cytometry. Neutrophils (CD45+ CD11b+ Ly6G+high; C–E) and neutrophils with intracellular macrophage markers (CD45+ CD11b+ Ly6G+high CD68+ F4/80+; F) were identified by flow cytometry. All gating was performed using FlowJo. All data are representative of two. All error bars represent SE. P-values were determined by a two-tailed Student’s t test (G–I) or two-way ANOVA (D–E); *, P < 0.05.
Complement proteins such as C3b and C1q can be deposited on necrotic cells; although complement also deposits on apoptotic bodies, this typically seems to occur after they undergo secondary necrosis (Colonna et al., 2016). The other complement fragments, C3a and C5a, play a role in vasodilation, chemotaxis, and extravasation of neutrophils (Gullstrand et al., 2009). We thus investigated the role of complement in efferocytosis of PITs. Addition of serum containing active complement (not heat inactivated) dramatically increased engulfment of PITs in vitro (Fig. 6 H). C3 is the convergence point of the three complement activation pathways (Figueroa and Densen, 1991), and as such, C3-deficient mice lack both C3b opsonization and C3a/C5a-mediated chemotaxis. Upon induction of pyroptosis (FliCind) in WT mice, there was a significant increase in the number of neutrophils in the spleen compared with control mice (Fig. 7 C) that was not observed in Casp1−/−Casp11−/− mice (Fig. 7 D). Notably, mice deficient in C3 both failed to recruit neutrophils to the spleen (Fig. 7 E) and had reduced efferocytosis of PITs in vivo (Fig. 7 F). This indicates that complement primarily mediates recruitment of neutrophils toward the site of the PIT, but it remains likely that complement also deposits on PITs to opsonize these structures for efferocytosis. Finally, mice deficient in C3 showed a significant decrease, but not a complete defect, in the clearance of FliCind S. Typhimurium (Fig. 7 G), demonstrating the importance of complement-mediated recruitment of neutrophils to the spleen in clearing bacteria after pyroptosis.
Because both fucoidan-treated and C3−/− mice had a partial reduction in FliCind S. Typhimurium clearance, we combined these experimental perturbations. C3−/− mice treated with fucoidan were entirely defective for FliCind S. Typhimurium clearance (Fig. 7 H), indicating that the combined action of complement receptors and scavenger receptors are critical for efferocytosis of the PIT and associated bacteria.
DISCUSSION
Many pathogens replicate within the macrophage intracellular niche. If inflammasomes succeed in detecting the pathogen, pyroptosis effectively eliminates the protected intracellular niche. After cell death, the organelles, cytoskeleton, and intracellular bacteria remain within the ruptured but largely intact plasma membrane, in what we define as the PIT. The bacteria entrapped within the PIT are not killed, and are only killed after phagocytosis of the PIT and entrapped bacterium by a neutrophil.
PITs are conceptually similar to NETs. Both promote bacterial clearance by entrapping bacteria, preventing their dissemination. However, NETs target extracellular bacteria, whereas PITs entrap bacteria that occupy the intracellular niche. NETs are intrinsically bactericidal because they contain adhered antimicrobial peptides and enzymes (Kaplan and Radic, 2012). In contrast, PITs damage, but do not kill bacteria. Our data suggests that NETs and PITs act independently of each other, as NET-defective mice (Mpo−/− and Elane−/−) are fully competent to clear bacteria trapped in PITs, and NETosis is not inhibited by the pan-caspase inhibitor z-VAD-fmk (Remijsen et al., 2011). Similar to PITs, apoptotic bodies have been shown to entrap intracellular mycobacteria and promote their clearance (Martin et al., 2012). Hence, entrapment of pathogens within cellular debris to enhance clearance constitutes a general strategy used by the immune system.
PITs are initiated by the opening of discrete pores that drive pyroptosis, rapidly leading to swelling and then frank membrane ruptures. With the exception of these isolated ruptures, however, the plasma membrane remains largely intact; it does not simply disintegrate. Instead, it retains intracellular bacteria and organelles. This trapping by a relatively intact plasma membrane is essential to maintaining the bacterial association with the dead cell and enabling bacterial clearance. If the bacteria were instead released to the extracellular space, they might more readily infect another susceptible cell. Instead, the PIT sequesters bacteria from other neighboring cells, allowing neutrophils time to migrate toward and efferocytose the PIT and the associated bacteria.
The infection is ultimately resolved not by the PIT itself, but by a secondary phagocyte that must phagocytose and kill the bacterium. The process of phagocytosing apoptotic bodies or necrotic debris has been termed efferocytosis, from Latin effere, meaning to bury a corpse, referring to the process by which dead cells are removed by phagocytes. Here, we propose to extend the term efferocytosis to include the phagocytosis of PITs. However, we propose that apoptotic bodies trapping bacteria will be mechanistically different from pore-induced cell death trapping. The morphology of the apoptotic body is vastly different from that which is seen after pore-induced cell death. Further, the find-me and eat-me signals will be very different, as we have begun to elucidate here.
After entrapping the previously intracellular bacteria, the PIT is recognized by the secondary neutrophil, which efferocytoses the PIT. Although it is possible that neutrophils are recruited to the site of pyroptosis by bacterial ligands, our data suggests that the secondary phagocyte primarily responds to host-derived find-me and eat-me signals. Indeed, neutrophils swarm toward a lysed cell immediately in vivo (Lämmermann et al., 2013), and FlaTox-induced PITs are efferocytosed, even though there are no bacteria. This ensures that any pathogen that triggers pyroptosis would be cleared, regardless of the surface composition of the microbe. We have identified two cellular components that play a role in efferocytosis of the PIT: complement and SRs. The PIT likely presents a smorgasbord of yet to be identified ligands for the SR-A family; SR-As are also proposed to participate in efferocytosis of apoptotic bodies and necrotic cell corpses (Peter et al., 2010). Exposure of high levels of PS on the cell surface of necrotic cells is known to be a ligand for C1q binding, thus driving C3b deposition (Païdassi et al., 2008). The role of complement is likely attributable to both complement-derived chemoattractants (C3a and C5a) and to complement-mediated opsonization (by C3b deposition). Given the ubiquitous presence of complement, and the redundancy seen with SRs, we propose that clearance of PIT-entrapped bacteria is extremely efficient.
Ultimately, the neutrophil must kill the bacteria. Among many antibacterial mechanisms, we have presented evidence that ROS generate by the phagocyte NADPH oxidase is critical (Miao et al., 2010a). Neutrophils are the primary source of ROS during bacterial infection, and thus PIT clearance was ablated by depletion of Ly6G+ cells in the S. Typhimurium FliCON model. However, macrophages also generate ROS, and thus we predict they could substitute for neutrophils when adequately stimulated depending on the cytokine milieu triggered by the specific pathogen. This may explain different results when L. monocytogenes is engineered to persistently express flagellin (Sauer et al., 2011). Likewise, non-ROS neutrophil/macrophage killing could be sufficient against some bacteria. It is therefore notable that PIT-associated bacteria are rendered more susceptible to killing not only by ROS, but also by antimicrobial peptides and antibiotics. The mechanism by which the bacteria are damaged, however, remains unclear. One possible hypothesis is that during pyroptosis the intracellular pressure increases as the cell swells, and then rapidly normalizes when the membrane ruptures. This rapid pressure change could damage the bacteria. Another possible mechanism is that mitochondrial ROS generated during the process of pyroptosis damages the bacteria within the PIT. However, we emphasize that the process of pyroptosis, although damaging to bacteria, was insufficient to kill them. We therefore consider the most important functions of the PIT are to trap the bacteria and promote efferocytosis.
It is important to note that although we have used S. Typhimurium as a tool with which to study and characterize the PIT, WT bacteria efficiently evade inflammasomes, pyroptosis, and the downstream consequences of entrapment in PITs (Miao and Rajan, 2011). We generated a S. Typhimurium strain that permitted tight temporal control of pyroptosis in the infected host cell. Without this type of experimental control, it would be very difficult to characterize the PIT and the ensuing immune response. However, because S. Typhimurium, L. monocytogenes, and many other vertebrate-adapted pathogens evade inflammasomes, during what infections will the PIT likely make a critical contribution to benefit host defense? B. thailandensis and Chromobacterium violaceum are nonvertebrate-adapted opportunistic pathogens; inflammasomes fully counteract their extreme virulence in vivo, with strong evidence pointing toward pyroptosis as a primary defense mechanism (Aachoui et al., 2015; Maltez et al., 2015). The PIT is likely to be an important mechanism to immobilize these bacteria after pyroptosis. Monitoring pyroptosis and PIT formation in vivo during infection by these bona fide inflammasome-activating bacteria will require the generation of different experimental approaches and tools.
Is the PIT a structure that is designed for this immunological purpose, or is it simply a passive byproduct of programmed cell death by osmotic swelling? We propose that the PIT is an inevitable consequence of cellular lysis via osmotic swelling, but that the innate immune system is specifically organized to take maximum advantage of this structure. Indeed, it is interesting to speculate that the genesis of cytosolic DAMPs originates from the need to drive neutrophil chemotaxis to the PIT.
Our data shows that pyroptosis initiates a multifaceted, coordinated, and protective innate immune response against intracellular pathogens. Pyroptosis results in the generation of a structure that entraps the previously intracellular bacteria, the PIT. The PIT drives clearance of intracellular bacteria after inflammasome detection, by dually containing the bacteria and elaborating signals that promote efferocytosis.
MATERIALS AND METHODS
Mice, mouse infections, and treatments
Wild type C57BL/6 (The Jackson Laboratory), Csf1r-GFP (The Jackson Laboratory), Ncf1−/− (The Jackson Laboratory), Casp1−/−Casp11−/− (Kuida et al., 1995), Mpo−/− (provided by Y. Aratani, Yokohama City University, Tokyo, Japan), Elane−/− (The Jackson Laboratory), C3−/− (The Jackson Laboratory), and Mertk−/− (G. Matsushima, University of North Carolina [UNC], Chapel Hill, NC) mice were housed in specific pathogen–free facility. All animal protocols were approved by the Institutional Animal Care and Use Committee at UNC Chapel Hill and met guidelines of the National Institutes of Health for the humane care of animals. For competitive indices, 8–10-wk-old mice were infected with 1,000 CFUs of a 1:1 ratio of stationary phase flgB::Tn10 (with kanamycin-resistant control vector; pWSK129) and flgB::Tn10 FliCind (ampicillin-resistant inducible flagellin plasmid; pEM087) S. Typhimurium in PBS by i.p. injection. At 17 h after infection, mice were treated with 0.8 mg of doxycycline to induce expression of fliC and 10 mg/kg fucoidan (Sigma-Aldrich), or 2 mg/kg 2-ThioUTP (Santa Cruz Biotechnology). Spleens, liver, and MLNs were harvested at 24 h after injection (7 h after doxycycline injection) and homogenized, and dilutions were plated onto LB + antibiotics. Competitive index are log flgB::Tn10 pFliCind CFU/flgB::Tn10 pWSK129 CFU. For neutrophil depletion, C57BL/6 mice were injected with 500 µg anti-Ly6G clone 1A8 (BioXCell) or isotype control IgG2a (BioXCell) at 12 h before infection.
Macrophage culture, bacterial strains, and infection
BMMs were isolated as previously described (Miao et al., 2010a). BMMs were stimulated with 50 ng/ml LPS overnight (only Fig. 1, E, F, and K), and infected by centrifugation at 250 g for 5 min. At 30 min after infection, cells were washed with PBS and replaced with media containing 15 µg/ml gentamicin (Sigma-Aldrich) and analyzed at the indicated total time points (i.e., 2.5-h infection is a 30 min infection followed by 2-h incubation with gentamicin). MOIs were chosen for each species to either minimize inflammasome activity or, only in the case of SPI1-induced S. Typhimurium, to maximize detection (detailed in Tables S1 and S2). For strains and culture conditions, see Tables S1 and S2. Supernatants were collected at the indicated time points. Cell lysis was determined by measuring LDH release (Rayamajhi et al., 2013b).
Live cell and confocal microscopy
For live and confocal microscopy, WT, Csf1r-GFP, and Casp1−/−Casp11−/− BMM cells were seeded in a MatTek dish or on regular coverslips, and LPS treated, infected, and gentamicin treated (15 µg/ml) as described above, or treated with 3 µg/ml FlaTox and 2 µM Nocodazole or 0.5 mg/ml cytochalasin D, and then fixed with 3% formaldehyde at 2 h after infection for confocal microscopy. Live cells were stained with 1 µM CellTracker Blue CMAC, 0.5 µM CellTracker Green CMFDA, 1 µM CellTracker Orange CMRA, 1 µM MitoTracker Green FM, 2.5 µM Bodipy-Green, 0.5 µM ER-Tracker Green, and 2.5 µg/ml WGA-Alexa Fluor 488, 10 nM PI, and 1 mg/ml Dextran-Alexa Fluor 555 (Invitrogen). Fixed cells were permeabilized, blocked in 5% BSA/PBS, and stained with 0.1 µM Alexa Fluor 555 Phalloidin (Invitrogen) or antivimentin or antitubulin antibodies, 2 mM DAPI (Sigma-Aldrich), or anti–caspase-1 (Genentech). For live cell microscopy, cells were imaged in a humidified chamber at 5% CO2 37°C starting 15 min after infection and for 2 h in phenol-free imaging media. All live cell and confocal images were acquired at 40× using a FV1200 SP5 confocal microscope (Olympus) and analyzed by Fluoview software (Olympus).
EM
WT BMMs were infected with L. monocytogenes multiplicity of infection (MOI) 20 or stationary or SPI1-induced S. Typhimurium MOI 50, treated with 3 µg/ml FlaTox, and fixed with 2.5% gluteraldehyde. Cells were treated with 1% osmium tetroxide and dehydrated with increasing concentrations of ethanol, infiltrated, and embedded in Polybed 812 epoxy resin. Monolayers were sectioned en face to the substrate at 70 nm using an ultramicrotome (Ultracut UCT; Leica) and diamond knife. Ultrathin sections collected on 200 mesh copper grids were post-stained with 4% aqueous uranyl acetate and lead citrate. Samples were imaged using a LEO EM910 transmission electron microscope at 80 kV (Carl Zeiss). Images were acquired using a Gatan Orius SC1000 CCD digital camera with Digital Micrograph 3.11.0 software.
Quantitation of cellular markers
WT BMMs were labeled, infected, or treated with 3 µg/ml FlaTox, 50 ng/ml TNF/10 µM ZVAD/5 µM BV6, 0.05% Saponin, or 1 µM staurosporine and subjected to live cell microscopy as described above. Mean fluorescence pixel intensity of ∼100 individual live or dead cells was quantitated using ImageJ software (National Institutes of Health). Dead cells were identified by either PI-staining or differential interference contrast. To assess the number of dead cells that remained attached to the surface of the imaging dish, three fields of ∼50 PI-negative GFP-BMMs versus PI-positive cells, were counted at start of imaging and at the end of video in two individual experiments.
In vitro phagocytosis of PITs
CellTracker Green–labeled WT BMMs were infected with SPI1-induced S. Typhimurium MOI 25. CellTracker Blue-labeled Casp1−/−Casp11−/− BMMs were added to the MatTek dish with PI. Alternatively, BMMs were treated with 3 µg/ml FlaTox and 1 µM fucoidan (Sigma-Aldrich) or 10 µM 2-ThioUTP tetrasodium salt (Santa Cruz Biotechnology, Inc.). At 2 h after infection, 10 fields of ∼50 live cells each were acquired. The rate of phagocytosis by Casp1−/−Casp11−/− BMMs was calculated by determining the percentage of blue BMM containing green pyroptotic debris and/or PI-positive nuclei.
In vitro bacterial viability and association assay
WT BMMs were infected and treated with gentamicin, as described above, and then treated with 3 µg/ml FlaTox, 50ng/ml TNF/10 µM ZVAD/5 µM BV6, 0.05% saponin, or 1 µM staurosporine for 2 h. To determine the number of GFP-bacteria associated with the PIT, infected cells were imaged by confocal microscopy. The number of GFP-bacteria associated with live and dead cells was manually counted for 10 fields (∼200 cells). Cells were lysed with Tx100 and plated on Luria broth.
In vitro H2O2, polymyxin B, ciprofloxacin susceptibility, and reinfection assay
WT BMMs were infected and treated with gentamicin as described above, and treated with PBS or 3 µg/ml FlaTox. Cells were lysed with Tx100 and cell lysates were added to 1 ml LB + H2O2, polymyxin B, or ciprofloxacin at 37°C for 2 h, and then plated on LB agar. To assess infection of bacteria associated with a live cell versus a PIT, cells were scraped off the plate and lysed by forcefully passaging supernatant through a 30-gauge needle three times. Infected cell lysates were added to Casp1−/−Casp11−/− BMMs for 20 min, washed, treated with 15 µg/ml gentamicin for 2 h, and fixed. Images were acquired by confocal microscopy, and the number of GFP-bacteria per cell was counted for ∼10 fields (∼200 cells).
Cell isolation and flow cytometry
For IP injection of PITs, 2 × 106 CellTracker Orange–labeled WT BMM were treated with 3 µg/ml FlaTox and IP injected into WT mice. Peritoneal fluid was collected at 2 h after IP injection and stained. For isolation of splenocytes, the spleen were treated with Collagenase D, RBC lysis buffer, and flushed through a cell strainer (Falcon). Cells were Fc-blocked with anti-CD16/CD32 and stained with anti–F4-80-PacBlue (BM8), CD11b-APC (M1/70), Ly6G-APC Cy7 (1A8), CD68-PE (FA-11) and Ly6C-FITC (AL-21) and cell viability marker Aqua dead live-AmCyan (all from BD), fixed, and analyzed on a LRSIII (BD; UNC Flow Cytometry Core Facility).
Amnis ImageStream
Single cells were isolated from spleen. Neutrophils were isolated from whole splenocytes using an anti-Ly6G (1A8+) MicroBead kit on an autoMACS column (Miltenyi Biotec). Isolated Ly6G+ cells were washed, fixed, and stained with anti–F4-80-PacBlue (BM8), Ly6G-APC Cy7 (1A8), and CD68-PE (FA-11; all from BD), and fixed again. Cells were analyzed on an Amnis ImageStreamX MarkII (UNC Flow Cytometry Core Facility).
Statistical analysis
All analysis and graphical presentations were made using Prism. All error bars represent SE. P-values were determined by a two-tailed Student’s t test.
Online supplemental material
Figure S1 shows spleens from mice infected IP with control or FliCind GFP–S. Typhimurium. Video 1 shows soluble GFP protein is released during pyroptosis (related to Fig. 1). Video 2 show bacteria are trapped in the pyroptotic cell corpse (related to Fig. 2). Video 3 shows macrophages phagocytose pyroptotic cell debris (related to Fig. 2). Video 4 shows macrophages phagocytose the entire pyroptotic cell corpse and trapped bacteria (related to Fig. 4). Video 5 shows multiple macrophages phagocytose a piece of the pyroptotic cell corpse each (related to Fig. 4). Tables S1 and S2 show bacterial strains, plasmids, and growth conditions. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151613/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dat Mao, Taylor Atherton, Davis Trihn, Cody Vientos, Jon Hagar, and Manira Rayamajhi for technical assistance and Glen Matsushima (UNC) for sharing mice.
This work was funded by National Institutes of Health grants DK007737 (I. Jorgensen), AI097518 (E.A. Miao), AI057141 (E.A. Miao), AI119073 (E.A. Miao), and AI077703 (B.A. Krantz). Research reported in this publication was supported by the Office of the Director, National Institutes of Health under award number 1 S10 OD017984-01A1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The UNC Flow Cytometry Core Facility is supported in part by P30 CA016086 Cancer Center Core Support grant to the UNC Lineberger Comprehensive Cancer Center.
The authors declare no competing financial interests.
Author contributions: I. Jorgensen and Y. Zhang performed the experiments. I. Jorgensen and E.A. Miao conceived, designed and analyzed the experiments, and wrote the paper. B.A. Krantz provided essential reagents.
Footnotes
Abbreviations used:
- BMM
- BM-derived macrophage
- EM
- electron microscopy
- FliCind
- inducible flagellin
- LDH
- lactate dehydrogenase
- MOI
- multiplicity of infection
- NAIP
- neuronal apoptosis inhibitory protein
- NET
- neutrophil extracellular trap
- PI
- propidium iodide
- PIT
- pore-induced intracellular trap
- SR
- scavenger receptor
- WGA
- wheat germ agglutinin
References
- Aachoui Y., Leaf I.A., Hagar J.A., Fontana M.F., Campos C.G., Zak D.E., Tan M.H., Cotter P.A., Vance R.E., Aderem A., and Miao E.A.. 2013. Caspase-11 protects against bacteria that escape the vacuole. Science. 339:975–978. 10.1126/science.1230751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aachoui Y., Kajiwara Y., Leaf I.A., Mao D., Ting J.P., Coers J., Aderem A., Buxbaum J.D., and Miao E.A.. 2015. Canonical inflammasomes drive IFN-γ to prime caspase-11 in defense against a cytosol-invasive bacterium. Cell Host Microbe. 18:320–332. 10.1016/j.chom.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan M.A., and Cookson B.T.. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38:31–40. 10.1046/j.1365-2958.2000.02103.x [DOI] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., and Zychlinsky A.. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Canton J., Neculai D., and Grinstein S.. 2013. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 13:621–634. 10.1038/nri3515 [DOI] [PubMed] [Google Scholar]
- Carvalho A.C., Sousa R.B., Franco A.X., Costa J.V., Neves L.M., Ribeiro R.A., Sutton R., Criddle D.N., Soares P.M., and de Souza M.H.. 2014. Protective effects of fucoidan, a P- and L-selectin inhibitor, in murine acute pancreatitis. Pancreas. 43:82–87. 10.1097/MPA.0b013e3182a63b9d [DOI] [PubMed] [Google Scholar]
- Colonna L., Parry G.C., Panicker S., and Elkon K.B.. 2016. Uncoupling complement C1s activation from C1q binding in apoptotic cell phagocytosis and immunosuppressive capacity. Clin. Immunol. 163:84–90. 10.1016/j.clim.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson B.T., and Brennan M.A.. 2001. Pro-inflammatory programmed cell death. Trends Microbiol. 9:113–114. 10.1016/S0966-842X(00)01936-3 [DOI] [PubMed] [Google Scholar]
- Elmore S. 2007. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35:495–516. 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa J.E., and Densen P.. 1991. Infectious diseases associated with complement deficiencies. Clin. Microbiol. Rev. 4:359–395. 10.1128/CMR.4.3.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S.L., and Cookson B.T.. 2006. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell. Microbiol. 8:1812–1825. 10.1111/j.1462-5822.2006.00751.x [DOI] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in Salmonella-infected macrophages. Nat. Immunol. 7:576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Gullstrand B., Mårtensson U., Sturfelt G., Bengtsson A.A., and Truedsson L.. 2009. Complement classical pathway components are all important in clearance of apoptotic and secondary necrotic cells. Clin. Exp. Immunol. 156:303–311. 10.1111/j.1365-2249.2009.03896.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I., and Miao E.A.. 2015. Pyroptotic cell death defends against intracellular pathogens. Immunol. Rev. 265:130–142. 10.1111/imr.12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M.J., and Radic M.. 2012. Neutrophil extracellular traps: double-edged swords of innate immunity. J. Immunol. 189:2689–2695. 10.4049/jimmunol.1201719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595. 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E., Jenne C.N., Surewaard B.G., Thanabalasuriar A., Lee W.Y., Sanz M.J., Mowen K., Opdenakker G., and Kubes P.. 2015. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat. Commun. 6:6673 10.1038/ncomms7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E.S., Baehrecke E.H., Blagosklonny M.V., El-Deiry W.S., Golstein P., Green D.R., et al. Nomenclature Committee on Cell Death 2009 . 2009. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16:3–11. 10.1038/cdd.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K., Lippke J.A., Ku G., Harding M.W., Livingston D.J., Su M.S., and Flavell R.A.. 1995. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 267:2000–2003. 10.1126/science.7535475 [DOI] [PubMed] [Google Scholar]
- Lämmermann T., Afonso P.V., Angermann B.R., Wang J.M., Kastenmüller W., Parent C.A., and Germain R.N.. 2013. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature. 498:371–375. 10.1038/nature12175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltez V.I., Tubbs A.L., Cook K.D., Aachoui Y., Falcone E.L., Holland S.M., Whitmire J.K., and Miao E.A.. 2015. Inflammasomes coordinate pyroptosis and natural killer cell cytotoxicity to clear infection by a ubiquitous environmental bacterium. Immunity. 43:987–997. 10.1016/j.immuni.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.J., Booty M.G., Rosebrock T.R., Nunes-Alves C., Desjardins D.M., Keren I., Fortune S.M., Remold H.G., and Behar S.M.. 2012. Efferocytosis is an innate antibacterial mechanism. Cell Host Microbe. 12:289–300. 10.1016/j.chom.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler K.D., Fuchs T.A., Nauseef W.M., Reumaux D., Roesler J., Schulze I., Wahn V., Papayannopoulos V., and Zychlinsky A.. 2011. Myeloperoxidase is required for neutrophil extracellular trap formation: implications for innate immunity. Blood. 117:953–959. 10.1182/blood-2010-06-290171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., and Rajan J.V.. 2011. Salmonella and caspase-1: A complex interplay of detection and evasion. Front. Microbiol. 2:85 10.3389/fmicb.2011.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., and Aderem A.. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569–575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Miao E.A., Leaf I.A., Treuting P.M., Mao D.P., Dors M., Sarkar A., Warren S.E., Wewers M.D., and Aderem A.. 2010a Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11:1136–1142. 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Mao D.P., Yudkovsky N., Bonneau R., Lorang C.G., Warren S.E., Leaf I.A., and Aderem A.. 2010b Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 107:3076–3080. 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E.S., Guo H., and Kaiser W.J.. 2015. Necroptosis: The Trojan horse in cell autonomous antiviral host defense. Virology. 479-480:160–166. 10.1016/j.virol.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J.E., Tedbury P.R., Homer-Vanniasinkam S., Walker J.H., and Ponnambalam S.. 2005. Biochemistry and cell biology of mammalian scavenger receptors. Atherosclerosis. 182:1–15. 10.1016/j.atherosclerosis.2005.03.036 [DOI] [PubMed] [Google Scholar]
- Païdassi H., Tacnet-Delorme P., Garlatti V., Darnault C., Ghebrehiwet B., Gaboriaud C., Arlaud G.J., and Frachet P.. 2008. C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J. Immunol. 180:2329–2338. 10.4049/jimmunol.180.4.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C., Wesselborg S., Herrmann M., and Lauber K.. 2010. Dangerous attraction: phagocyte recruitment and danger signals of apoptotic and necrotic cells. Apoptosis. 15:1007–1028. 10.1007/s10495-010-0472-1 [DOI] [PubMed] [Google Scholar]
- Poon I.K., Hulett M.D., and Parish C.R.. 2010. Molecular mechanisms of late apoptotic/necrotic cell clearance. Cell Death Differ. 17:381–397. 10.1038/cdd.2009.195 [DOI] [PubMed] [Google Scholar]
- Ravichandran K.S. 2011. Beginnings of a good apoptotic meal: the find-me and eat-me signaling pathways. Immunity. 35:445–455. 10.1016/j.immuni.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Zak D.E., Chavarria-Smith J., Vance R.E., and Miao E.A.. 2013a Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191:3986–3989. 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Zhang Y., and Miao E.A.. 2013b Detection of pyroptosis by measuring released lactate dehydrogenase activity. Methods Mol. Biol. 1040:85–90. 10.1007/978-1-62703-523-1_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remijsen Q., Vanden Berghe T., Wirawan E., Asselbergh B., Parthoens E., De Rycke R., Noppen S., Delforge M., Willems J., and Vandenabeele P.. 2011. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 21:290–304. 10.1038/cr.2010.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J.D., Pereyre S., Archer K.A., Burke T.P., Hanson B., Lauer P., and Portnoy D.A.. 2011. Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc. Natl. Acad. Sci. USA. 108:12419–12424. 10.1073/pnas.1019041108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Trinidad N.J., Moayeri M., Kintzer A.F., Wang S.B., van Rooijen N., Brown C.R., Krantz B.A., Leppla S.H., Gronert K., and Vance R.E.. 2012. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 490:107–111. 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- Zychlinsky A., Prevost M.C., and Sansonetti P.J.. 1992. Shigella flexneri induces apoptosis in infected macrophages. Nature. 358:167–169. 10.1038/358167a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.