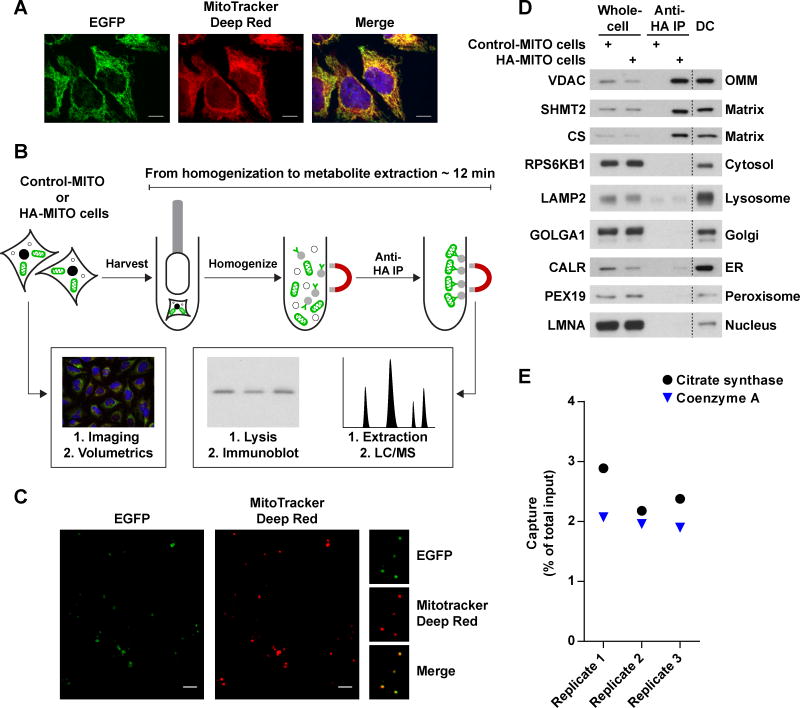
Figure 1. see also Figure S1: A method for the rapid and specific isolation of intact mitochondria.
(A) The 3XTag-EGFP-OMP25 protein properly localizes to mitochondria. Representative confocal micrographs of HeLa cells expressing the recombinant EGFP-fusion protein (green). Mitochondria and nuclei were stained with MitoTracker Deep Red FM (red) and Hoechst (blue), respectively. Scale bars, 10 μm.
(B) Workflow for the absolute quantification of matrix metabolites. Cells expressing Control-MITO (Control-MITO cells) or HA-MITO (HA-MITO cells) are rapidly harvested and dounce homogenized. HA-tagged mitochondria are isolated with a 3.5 minute IP, washed, and then lysed for immunoblot analysis to determine the amount of captured mitochondria or extracted for LC/MS-based metabolomics to quantify metabolites. Confocal microscopy and volumetric analysis of the HA-MITO-expressing cells are used to determine total mitochondrial volume per cell, which is then adjusted based on the percentage of mitochondrial volume occupied by the matrix (~63.16% of mitochondrial volume = matrix) (Gerencser et al., 2012). All of these measurements are combined to calculate the matrix concentration of a metabolite.
(C) Epitope-tagged mitochondria isolated from cells incubated with MitoTracker Deep Red FM retain the dye. Representative confocal micrographs of beads with isolated mitochondria (green) and MitoTracker Deep Red FM signal (red). On the right are magnifications of several beads with mitochondria. Scale bars, 5 μm.
(D) Purification of epitope-tagged mitochondria has significantly less organellar contamination compared to a differential centrifugation method optimized for speed. Immunoblot analysis of whole-cell lysates (Whole-cell) and lysates of mitochondria purified with anti-HA beads (Anti-HA IP) or differential centrifugation (DC). Lysates were derived from cells expressing Control-MITO (Control-MITO cells) or HA-MITO (HA-MITO cells). The names of the protein markers used are to the left of the blots and their corresponding subcellular compartments to the right. OMM, outer mitochondrial membrane; matrix, mitochondrial matrix; Golgi, Golgi complex; ER, endoplasmic reticulum.
(E) Epitope-tagged mitochondria retain both soluble proteins and small molecules to similar degrees. A comparison of the amount of captured mitochondria as assessed by a matrix protein (Citrate synthase) and a matrix metabolite (Coenzyme A). Data are represented as a percentage of the total material present in harvested cells. Cells were cultured in DMEM without pyruvate.