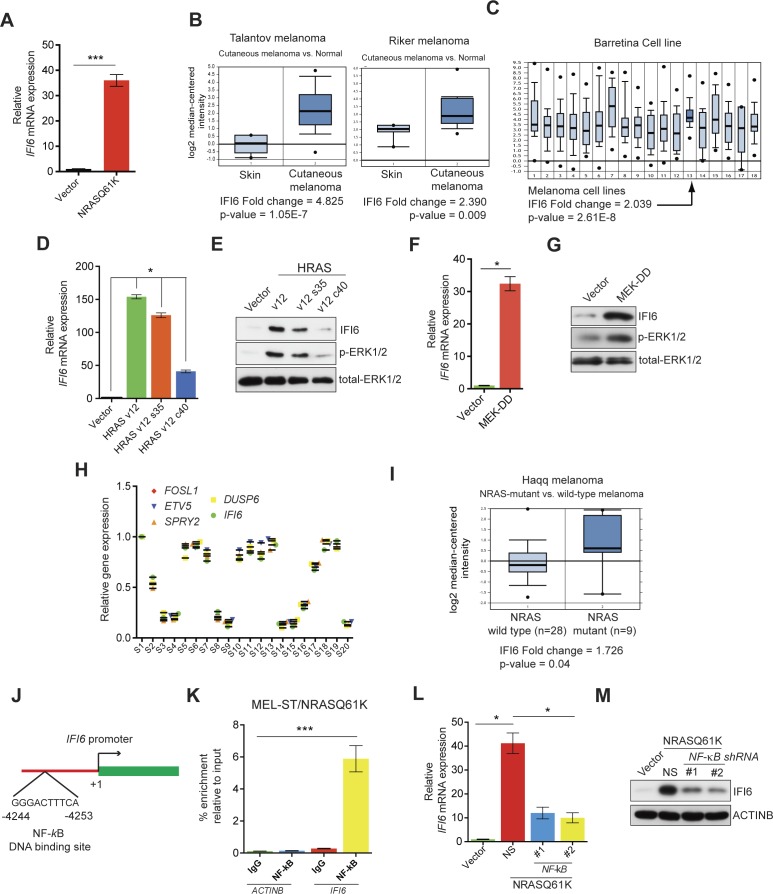
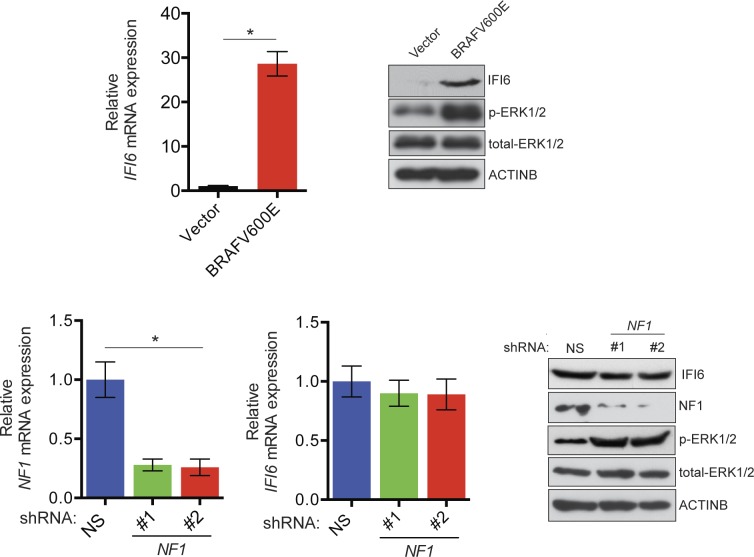
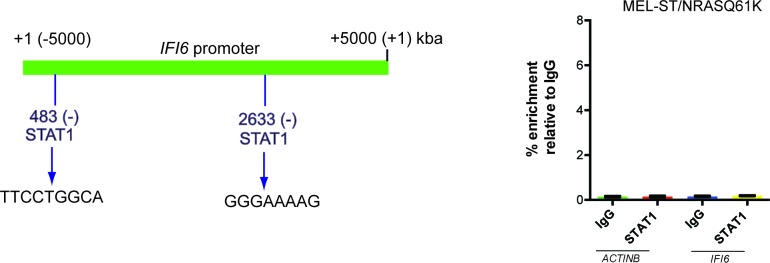
Figure 1. IFI6 is transcriptionally upregulated by NRASQ61K via MAPK pathway.
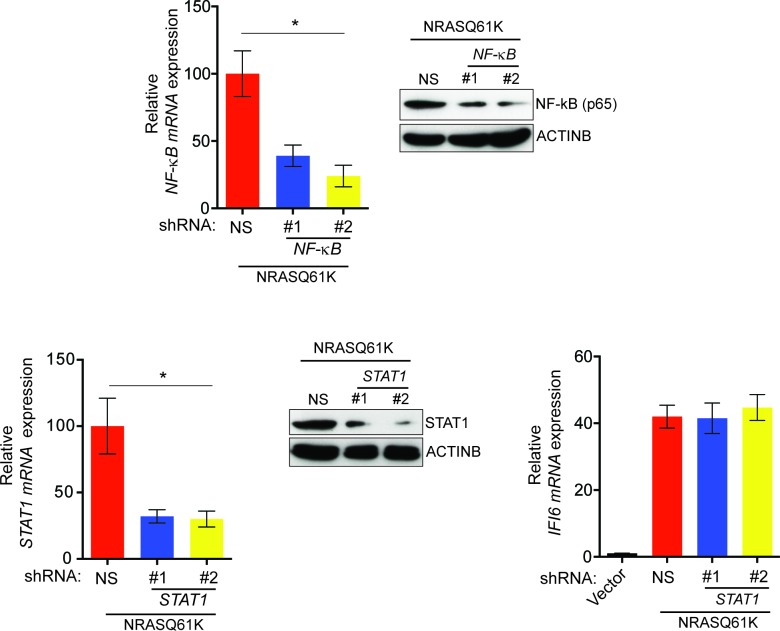
(A) Relative IFI6 mRNA expression in MEL-ST/NRASQ61K cells compared to empty vector-expressing MEL-ST cells. (B) Box plots for IFI6 mRNA expression in indicated melanoma gene expression datasets show significantly higher IFI6 mRNA expression in patient-derived cutaneous melanoma samples (2) compared to normal skin controls (1). (C) Box plot for IFI6 mRNA expression in Barretina cell line dataset. Lane 13 shows average of IFI6 mRNA expression in melanoma cell lines. (D) MEL-ST cells expressing empty vector or indicated HRAS mutants were analyzed by RT-qPCR. The relative expression of IFI6 mRNA in HRAS mutant-expressing MEL-ST cells compared to empty vector-expressing MEL-ST cells. (E) Relative expression of indicated proteins was evaluated by immunoblotting in MEL-ST cells expressing an empty vector or indicated HRAS mutants. (F) MEL-ST cells expressing empty vector or MEK-DD were analyzed for IFI6 mRNA expression by RT-qPCR. IFI6 mRNA expression in MEK-DD expressing MEL-ST cells relative to empty vector is shown. (G) MEL-ST cells expressing empty vector or MEK-DD were analyzed for indicated proteins by immunoblotting. (H) Analysis of patient-derived melanoma samples (n = 20) reveals co-expression of IFI6 with MAPK target genes. (I) Box plot for indicated melanoma gene expression dataset shows significantly higher IF6 mRNA expression in patient-derived NRAS-mutant melanoma samples (2) compared to NRAS wild-type melanoma samples (1). (J) Schematic presentation of NF-κB DNA binding site on the IFI6 promoter. (K) MEL-ST/NRASQ61K cells were analyzed for NF-κB enrichment using the ChIP assay.% NF-κB enrichment in comparison to IgG on the ACTIN or IFI6 gene promoter is shown. (L) MEL-ST cells expressing empty vector or NRASQ61K with NS or NF-κB shRNAs were analyzed for IFI6 mRNA expression by RT-qPCR. Relative IFI6 mRNA in comparison to empty vector expressing MEL-ST cells is shown. (M) MEL-ST cells expressing empty vector or NRASQ61K with NS or NF-κB shRNAs were analyzed for IFI6 protein levels by immunoblotting. ACTINB served as the loading control. In all panels, data are presented as mean ± SEM, and *p<0.05 and ***p<0.0005.
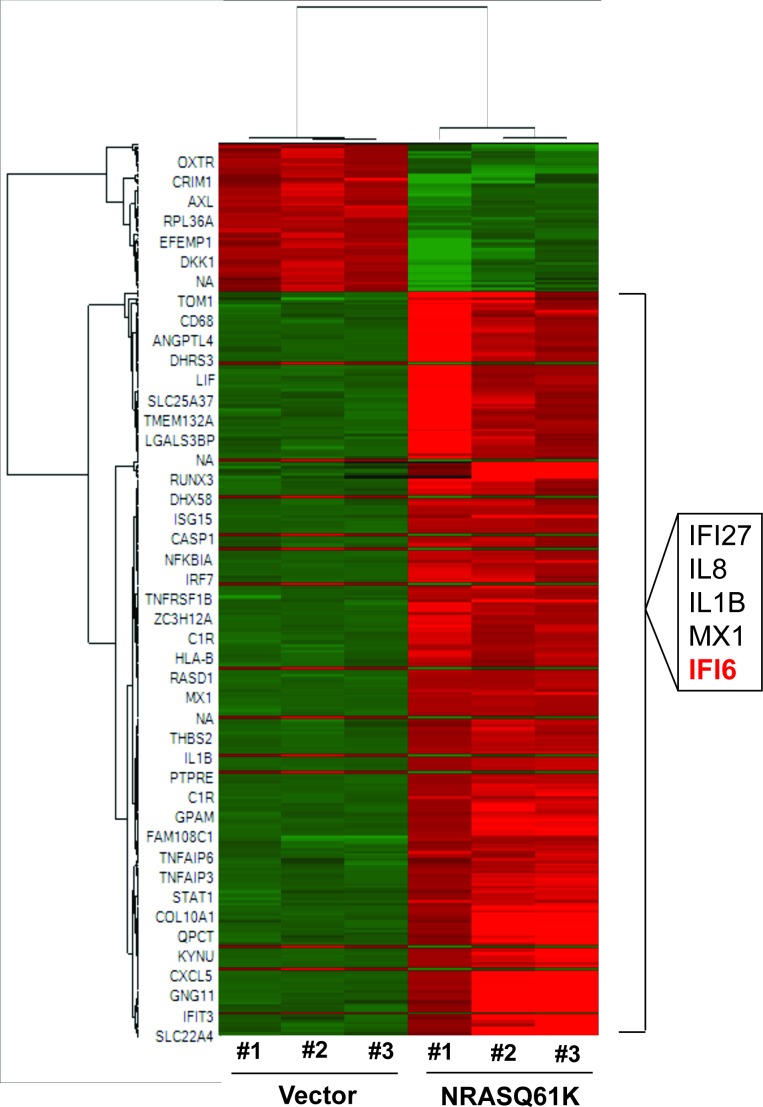
Figure 1—figure supplement 1. NRASQ61K transcriptionally upregulates interferon-stimulated genes.
Figure 1—figure supplement 2. Regulation of MX1 and IFI6 by NRASQ61K.