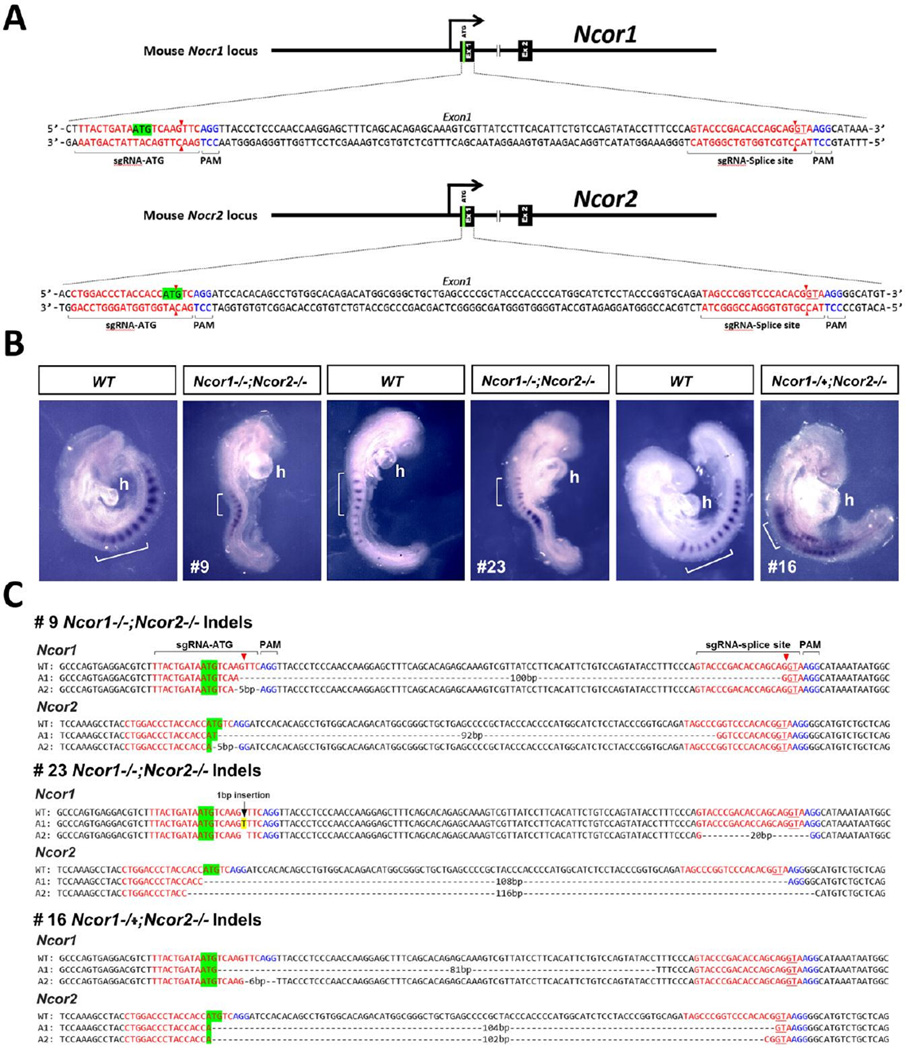
Fig. 1.
CRISPR/Cas9 gene editing of both Ncor1 and Ncor2 results in disruption of somitogenesis and heart development. (A) Strategy for generating mutations in the first exons of Ncor1 and Ncor2 using sgRNAs targeting the ATG start codon and GT splice donor site. (B) Shown is expression of the somite marker Uncx for wild-type (WT) or CRISPR mutants; brackets mark 6-somite regions for each embryo; h, heart (mutants exhibit distended heart tube). (C) DNA sequences for WT and both alleles (A1 and A2; sense strand) of Ncor1 and Ncor2 for the three CRISPR mutants shown above demonstrate that each mutant carries biallelic insertion or deletion mutations in both Ncor1 and Ncor2 that disrupt the ATG start codon, GT splice donor site, or reading frame. As Ncor1 A2 for embryo #16 carries an in-frame 6 bp deletion expected to delete only amino acids 4 and 5, this embryo is likely to have three null alleles rather than four.