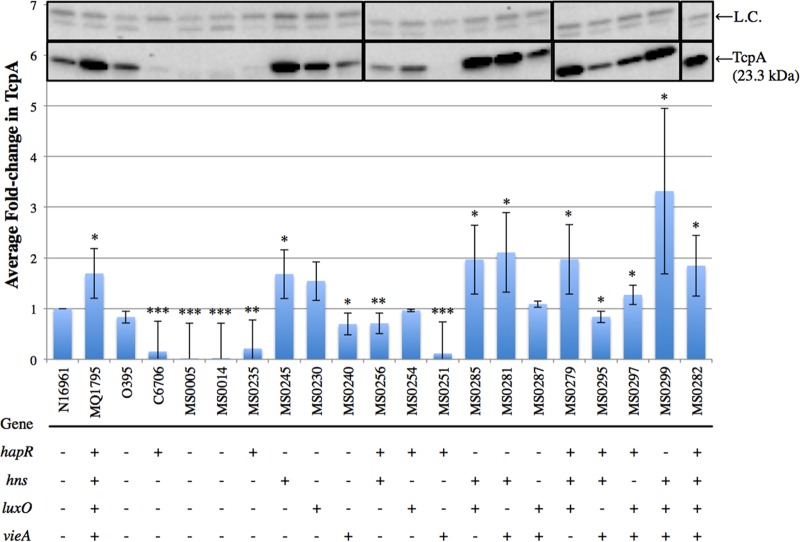
FIG 2 .
TcpA production profiles of single-SNP strains and double-, triple-, and quadruple-SNP combination strains. WCE were prepared, 3 µg of total protein was loaded onto a 16% Tris-glycine polyacrylamide gel, and proteins in immunoblots were quantified using densitometry for V. cholerae virulence factor TcpA. Immunoblots (inset) are from different blots (indicated by borders), all with WT N16961 and other controls run simultaneously. Nonspecific bands serve as a loading control (L.C.) and are labeled accordingly. Average fold changes were calculated relative to the TcpA level in N16961. Other controls included MQ1795 (hypervirulent clinical isolate), classical O395, El Tor C6706, O395ΔtoxT, and O395ΔtcpA. Strains containing the different SNPs and SNP combinations introduced into WT N16961 are indicated by the numbers beginning with “MS” found in Table 2. The presence of SNPs identified in genes in the indicated clinical isolates is denoted by a “+,” whereas the WT N16961 version of a SNP is denoted by a “−.” A two-tailed Student t test yielded P values of ≤0.05 (*), <0.005 (**), and <0.0005 (***).