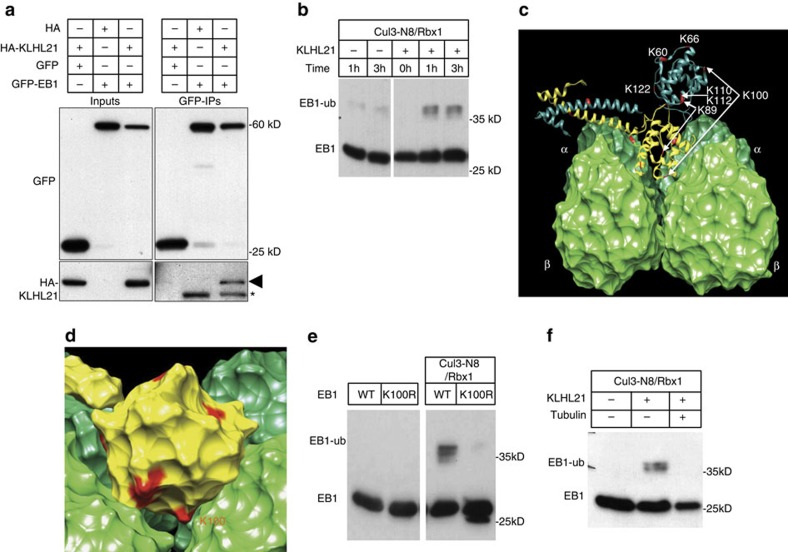
Figure 3. EB1 is preferentially ubiquitylated on lysine 100.
(a) HeLa cells were co-transfected as indicated with plasmids expressing HA-KLHL21 and GFP-EB1, or the corresponding empty HA- or GFP controls. Cell extracts were incubated with anti-GFP antibodies (GFP-IP) and bound proteins analysed by immunoblotting with anti-GFP (upper panels) or HA antibodies (lower panels). An aliquot of the cell extract was used to control protein expression (input). The black arrow marks HA-KLHL21 co-immunoprecipitating with GFP-EB1, while the asterisk points to GFP-EB1 cross reacting with the secondary antibody used for immunoblotting. (b) Purified EB1 was incubated with reconstituted and neddylated (N8) Cul3/Rbx1/UbH5 and ubiquitin (Ub) in the absence (−) or presence (+) of KLHL21 purified from E. coli. At the indicated times (hours), the reaction was stopped and analysed by immunoblotting with EB1-antibodies. The position of unmodified (EB1) and ubiquitylated EB1 (EB1-ub) is indicated. (c) Schematic representation of a EB1-dimer (yellow and cyan) binding to α-β tubulin (dark and light green, respectively, adapted from Maurer et al.11 and Slep et al.31). The positions of the ubiquitylated lysine residues identified by mass spectrometry from the in vitro reaction shown in d are indicated. (d) The position of lysine 100 (K100) was mapped on EB1 surfaces known to be involved in Mt binding. (e) Recombinant EB1 or EB1K100R alone (left panel) or with recombinant E2 and E3 enzymes (right panel) were subjected to in vitro ubiquitylation reactions as described in d. (f) Recombinant EB1 was subjected to in vitro ubiquitylation reactions with recombinant E2 and E3 enzymes and ubiquitin as described in d in the presence (+) or absence (−) of KLHL21 and stabilized Mts. The position of unmodified (EB1) and ubiquitylated EB1 (EB1-ub) is indicated.