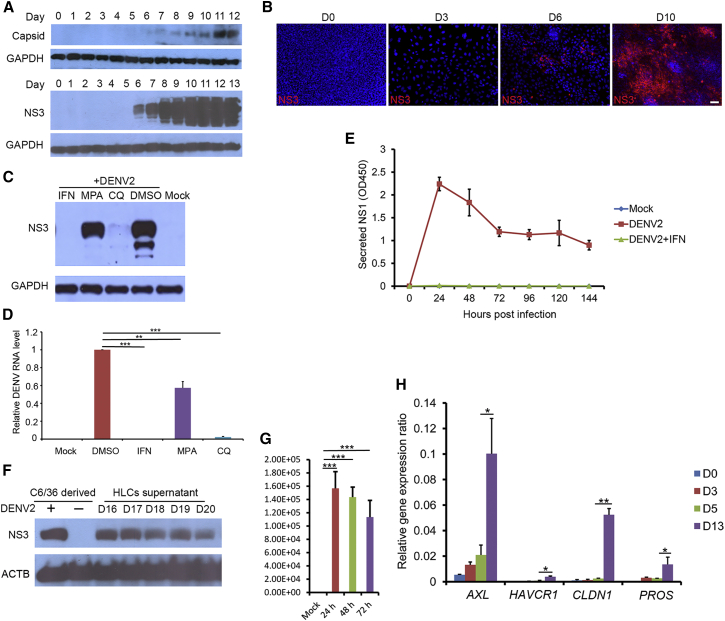
Figure 2.
HLCs Supported Persistent and Productive Infection of DENV
(A) DENV2 infection of HLCs in a differentiation stage-dependent manner. The hESCs (day 0) and differentiated cells at various differentiation stages from day 1 to day 13 were inoculated with DENV2 (strain 16681) at an MOI of 2 for 48 hr, and cell lysates were collected for western blotting.
(B) Immunostaining analysis of DENV2-infected HLCs. hESCs (day 0), definitive endoderm (day 3), hepatic progenitors (day 6), and HLCs (day 10) were infected by DENV2 as described above. Cell were fixed and stained with anti-NS3 antibody. Scale bar, 50 μm.
(C) DENV2 infection was blocked in HLCs by inhibitors. HLCs at day 15 after differentiation were pretreated with IFN-α2a (1,000 U/mL), MPA (1 μM), CQ (5 μg/mL), or DMSO (0.1%, v/v) for 8 hr, then subjected to DENV2 infection as described above.
(D) qRT-PCR analysis of DENV RNA levels. Cells were treated as described above. The relative DENV RNA level in each sample was normalized to DMSO-treated samples.
(E) Persistent replication of DENV2 in HLCs. Day-15 HLCs were exposed to DENV2 for 6 hr before the cells were stringently washed. NS1 protein in the supernatant collected every 24 hr after infection was measured by ELISA assay.
(F) DENV2 particles released from HLCs efficiently infected C6/36 cells. HLCs at day 15 were infected with DENV2 at an MOI of 2 for 6 hr. The inoculum was then removed and the cells washed stringently with PBS five times before the fresh medium was added. The supernatants were collected every 24 hr from day 16 to day 20. After each change of the spent medium, cells were washed stringently as described above. Each medium supernatant was added to naive C6/36 cells and incubated for 24 hr. Cells were then lysed and subjected to western blotting analysis. DENV2 derived from C6/36 cells was used as a positive control.
(G) Virus titration. Virus titers were calculated in the above supernatants collected at 24, 48, and 72 hr post infection.
(H) Upregulation of DENV entry factors during the differentiation process. Total RNA from differentiated cells at the indicated days were extracted and subjected to real-time qRT-PCR to measure the cellular expressions of AXL, HAVCR1, PROS, and CLDN1. GAPDH was used as the reference and its expression was set as 1. All the genes were normalized to GAPDH to obtain their relative gene-expression ratio.
Three independent experiments were performed. Data are presented as mean ± SD. p Values were calculated by Student's t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S2.