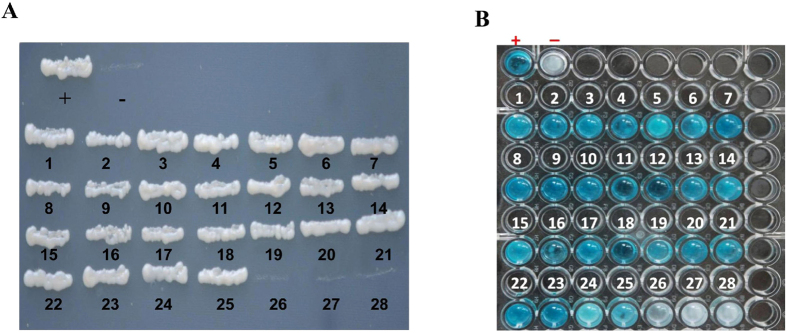
Figure 4.
Confirmation assays: (A) Retransformation analysis, (B) β-galactosidase assay. (A) Each of the 28 putative interactor proteins was used with SRBSDV-P10 protein to cotransform yeast strain NMY51 cells to confirm the interaction and eliminate false positives; pDHB1-LargeT + pDSL-p53 (positive control), pDHB1-LargeT + pPR3-N-empty prey insert (negative control). (B) β-galactosidase assay to identify the strength of interaction between SRBSDV-P10 protein and each of the 28 putative proteins. (+) = Positive control, (−) = Negative control. (1) NADH dehydrogenase subunit 1, (2) polyubiquitin, (3) alpha tubulin 84B, (4) growth hormone-inducible transmembrane protein, (5) longwave opsin, (6) cytochrome B, (7) sugar transporter 2, (8) vesicle-associated membrane protein 7, (9) sugar transporter 6, (10) ATP synthase lipid-binding protein, (11) nascent polypeptide-associated complex subunit alpha, (12) cuticlin-1, (13) ATP citrate lyase, (14) titin, (15) glucosyl glucuronosyl transferases, (16) tetraspanin 39D, (17) coronin-1C, (18) atlastin, (19) calcium-transporting ATPase sarcoplasmic/endoplasmic reticulum type, (20) integral membrane protein 2B, (21) integrin beta-3, (22) vesicle transport V-SNARE protein Vti1A, (23) tubulin beta-2C chain, (24) cytochrome C oxidase subunit III, (25) cathepsin L, (26) carboxylesterase, (27) conserved hypothetical protein, (28) vitellogenin.