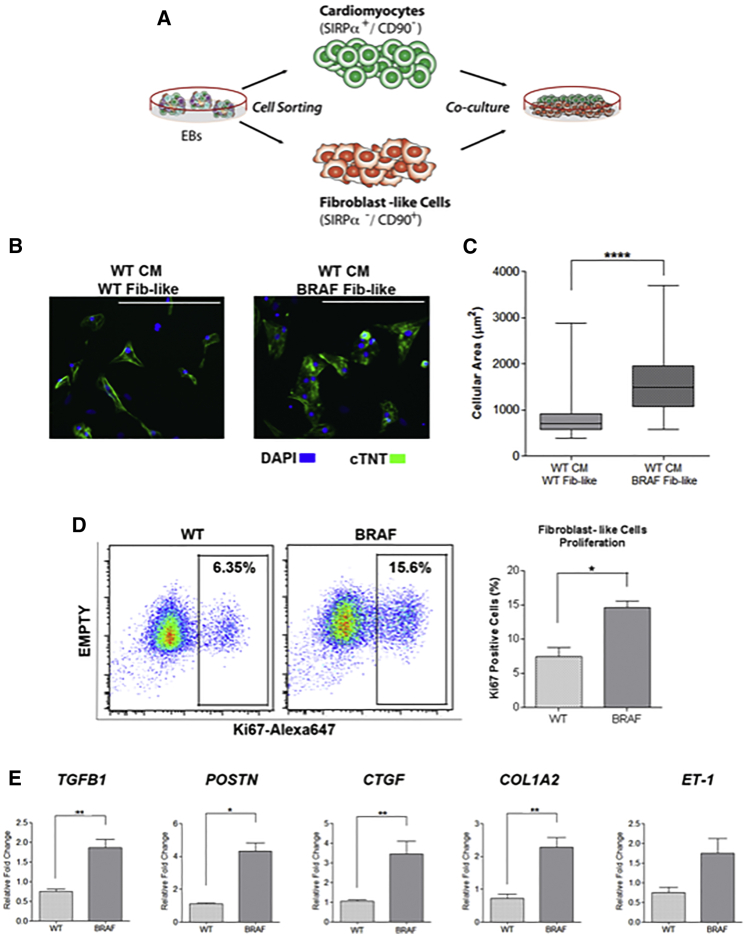
Figure 5.
BRAF-Mutant FLCs Influence the Hypertrophic Phenotype
(A) Schematic of co-culture experiment. WT CMs and WT and BRAF-mutant FLCs (Fib-like) were sorted from EBs and re-cultured together.
(B) Representative images of co-culture treatment groups stained with cTNT. Scale bars, 200 μm.
(C) Quantification of cellular area as depicted in (B). WT CMs became significantly larger upon co-culture with BRAF-mutant Fib-like cells (n = 67) compared with co-culture with WT Fib-like cells (n = 62) (∗∗∗∗p < 0.0001). Box-and-whisker plots show the median to the first and third quartiles and the minimum and maximum values. Data represent two biological (WT2, 3; BRAF1, 2) and three technical replicates.
(D) Increased proliferation rate of purified BRAF-mutant Fib-like cells (n = 3) compared with WT (n = 2) as demonstrated by increased staining for Ki67 by flow cytometry (∗p = 0.03). Data represent two (WT2, 3) or three (BRAF1, 2, 3) biological and two technical replicates. Data are presented as means ± SEM.
(E) Purified BRAF-mutant Fib-like cells expressed increased levels of fibrosis-associated genes compared with WT; TGFβ1 (BRAF n = 24, WT n = 18, ∗∗p = 0.002), POSTN (BRAF n = 15, WT n = 3, ∗p = 0.01), CTGF (BRAF n = 18, WT n = 12, ∗∗p = 0.002), COL1A2 (BRAF n = 24, WT n = 15, ∗∗p = 0.003), and ET-1 (BRAF n = 15, WT n = 8, p = 0.07, not significant). Data represent three biological (WT1, 2, 3; BRAF1, 2, 3) and three technical replicates. Data are presented as means ± SEM.