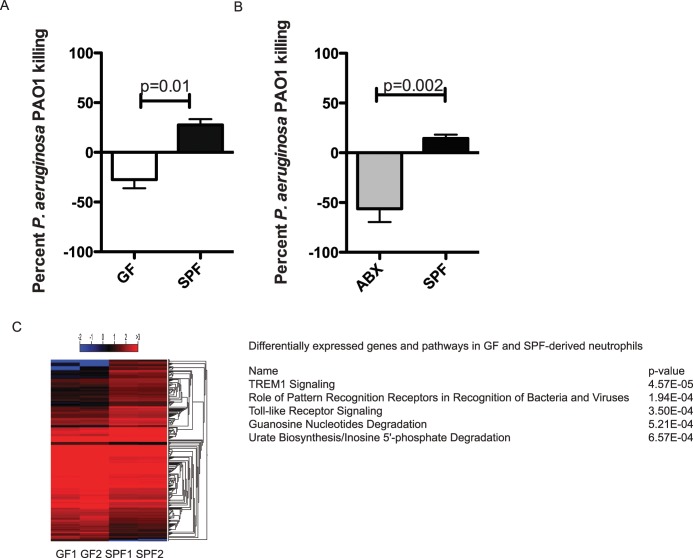
Fig 7. PMNs derived from either GF mice or mice treated with oral antibiotics have significantly decreased bactericidal activities against P. aeruginosa and altered transcriptomic signatures.
A. Percent P. aeruginosa PAO1 killing by GF PMNs and SPF SW-derived PMNs in vitro (p = 0.01, Unpaired Student t-test). Bone marrow-derived mature GF and SPF SW PMNs were exposed to P. aeruginosa PAO1 for 90 min at 37°C. Upon completion of incubation, an aliquot from the reaction mixture was plated to quantify the remaining PAO1. Neutrophils derived from animals without commensal microbiota display significantly reduced bactericidal capacity against P. aeruginosa. B. Percent of P. aeruginosa PAO1 killing by SPF SW-derived PMNs and PMNs purified from mice treated with oral antibiotic cocktail in vitro (p = 0.002, unpaired Student t-test). Neutrophils derived from mice that received oral antibiotics display significantly reduced bactericidal capacity against P. aeruginosa, illustrating that microbiota-derived signals promote neutrophil function. C. GF-derived neutrophils have significantly different transcriptional signature when compared to SPF-derived neutrophils. Shown are the transcriptomic profiles of differentially expressed genes in GF- and SPF-derived PMNs obtained by RNAseq analysis and the predicted pathways involved in PMN maturation in a heat map of the differentially expressed genes in GF- and SPF-derived PMNs. Hierarchical cluster analysis was performed on CLC Genomics workbench on subset of transcripts that show differential presence between the GF- and SPF-derived neutrophils (≥ 1.5-fold; p<0.05). The same subset was used to predict the potential upstream regulators by IPA to identify pathways that are involved in priming neutrophils for optimal function.