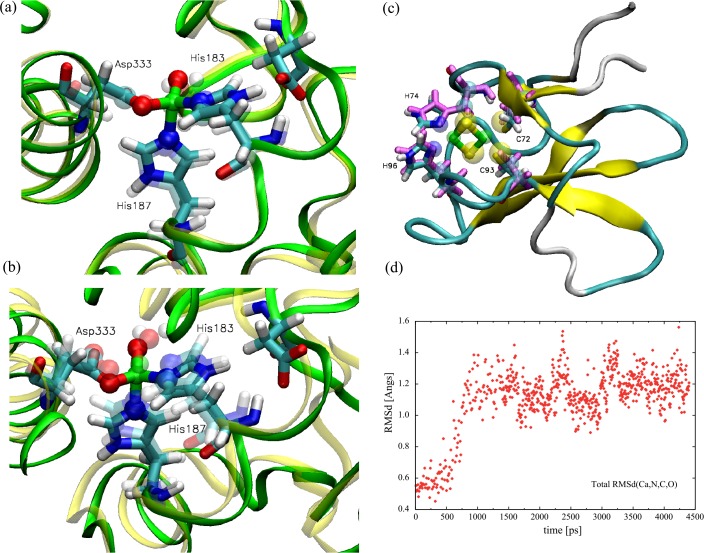
Fig 10. Snapshots taken after a short simulation time showing the variation of the structure from the original PDB file (id: 1WW9).
The simulations were carried out using using the force field parmeters developed in this work. (a) The subsequence of dOx comparing the minimized initial structure with the PDB structure (transparent residues). (b) The same substructure of 1WW9 after a short 4 ns MD simulation. Whereas the truncated structures tend to distort slightly during the simulation (as expected), this shows that the structure around the Fe center remains stable and relatively unchanged from the original PDB structure (transparent residues). (c) The same study for the Rieske region using a subsequence fragment of a related 1,9a-dioxygenase (PDB id: 3GKQ). The transparent residues are associated with the initial pdb data. (d) The total RMS deviation from the x-ray structure for a full MD simulation of the 3GKQ fragment using the developed force field parameters.