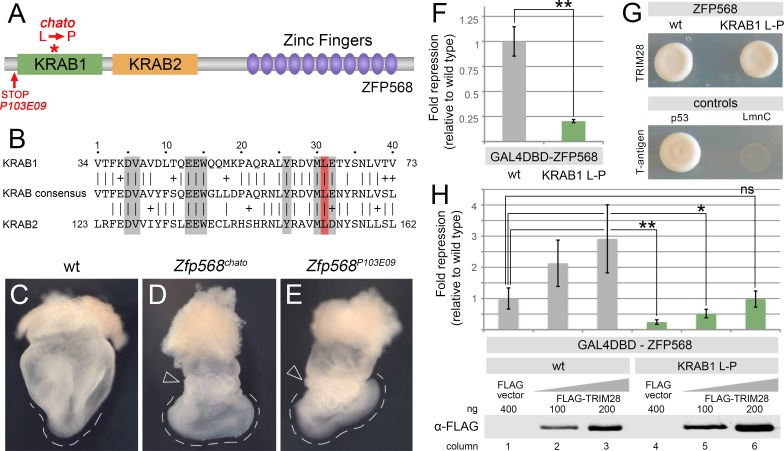
Fig 1. The chato mutation disrupts ZFP568 repressive activity, but not its ability to bind TRIM28.
(A) Domain structure of ZFP568. The red asterisk indicates the location of the chato mutation, which causes a Leu to Pro substitution. The red arrow indicates the position of the early stop codon in P103E09 mutants, which results in a truncated protein containing only the first ZFP568 11 amino acids. (B) Sequence alignment of the first and second KRAB motifs of ZFP568 with the Pfam KRAB consensus. Grey boxes indicate residues important for repressive activity as described in [11–13,25]. The red box highlights the conserved Leu residue mutated in chato. (C-E) Whole mount wild type (C), Zfp568chato (D) and Zfp568P103E09 (E) E8.5 embryos. The discontinued line highlights the abnormal U-shape of Zfp568 mutants, as compared with the V-shaped profile of wild type embryos. Empty arrowheads point to the yolk sac, which is abnormally ruffled in Zfp568 mutants. (F) Quantification of luciferase expression from a 5xUAS-luciferase reporter in HEK293T cells, in the presence of wild type (grey) and a KRAB1 L-P mutant (green) GAL4DBD-ZFP568. (G) Yeast two-hybrid assay for TRIM28 interaction with wild type ZFP568 (upper left) and a KRAB1 Leu-Pro mutant ZFP568 (upper right). T-antigen and p53 interaction was used as positive control. T-antigen and LmnC were used as negative control. (H) Quantification of luciferase expression from a 5xUAS-luciferase reporter in HEK293T cells, in the presence of wild type (grey) and KRAB1 Leu-Pro mutant (green) GAL4DBD-ZFP568, either in the absence (columns 1, 4) or presence of increasing amounts of FLAG-TRIM28 (columns 2–3 & 5–6). Luciferase expression is graphed as fold repression normalized to Gal4DBD (empty vector) and relative to wild type (column 1). Error bars represent standard deviation. Western blots show levels of FLAG-TRIM28 protein for each condition. Asterisks indicate samples which pairwise comparison had a p<0.05 (*) or p<0.005 (**). ns, no statistical significance.