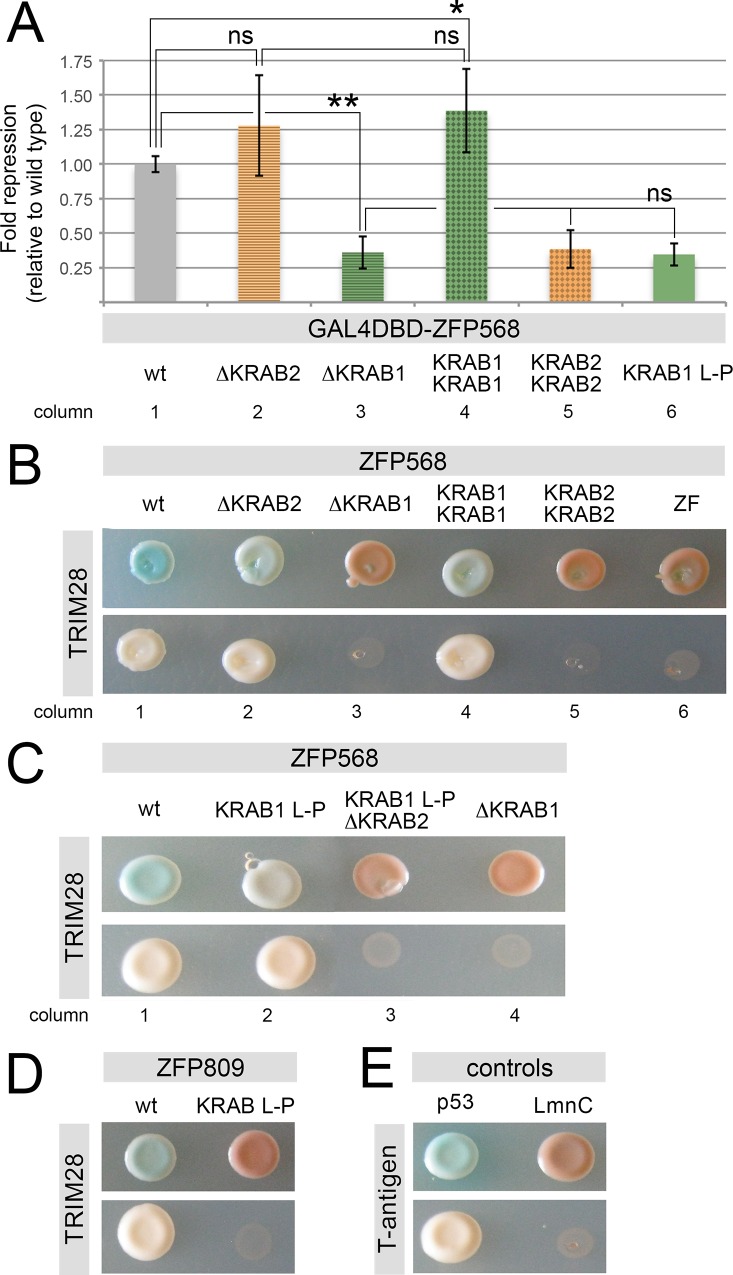
Fig 4. The repressive activity of ZFP568 depends on the properties of its KRAB motifs, but not on their number or location within the protein.
(A) Quantification of luciferase expression from a 5xUAS-luciferase reporter in HEK293T cells, in the presence of wild type (grey) and mutant versions of GAL4DBD-ZFP568: lacking the second KRAB motif (∆KRAB2), lacking the first KRAB motif (∆KRAB1), with two KRAB1 motifs (KRAB1 KRAB1), with two KRAB2 motifs (KRAB2 KRAB2), or with a mutated KRAB1 motif (KRAB1 L-P). Luciferase expression is graphed as fold repression compared to Gal4DBD (empty vector) and relative to wild type (column 1). Error bars represent standard deviation. (B-D) Yeast two-hybrid assays showing the interaction of TRIM28 with wild type and mutant versions of ZFP568 and ZFP809 as indicated. (E) T-antigen and p53 were used as positive controls. T-antigen and LmnC were used as negative controls. A truncated ZFP568 lacking all KRAB motifs (ZF) was also used as a negative control in panel B (column 6). The pictures show representative yeast colonies for each of the interactions tested, as grown on low stringency media (upper row) and high stringency media (lower row). Asterisks indicate samples which pairwise comparison had a p<0.05 (*) or p<0.005 (**). ns, no statistical significance.