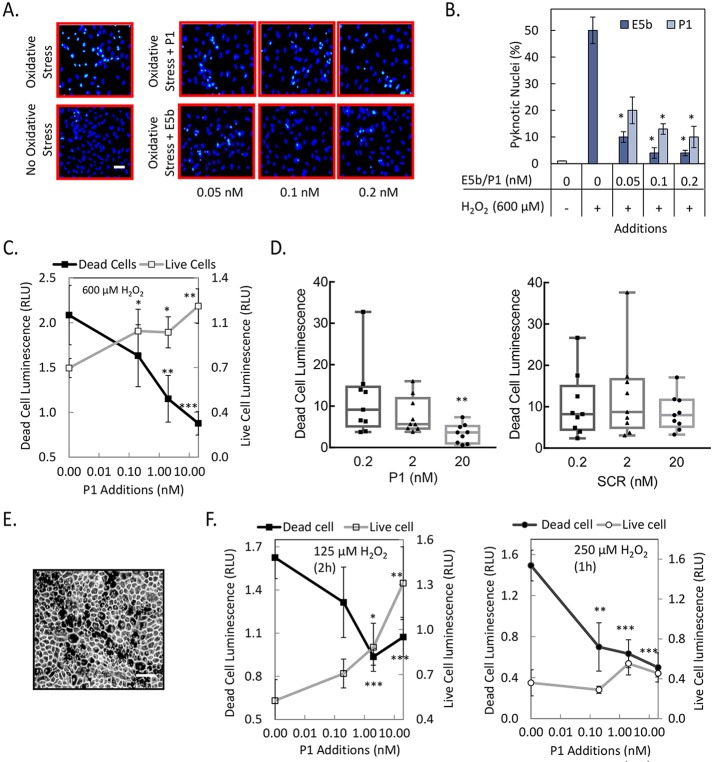
Figure 4.
Oxidative stress in ARPE-19 and pig primary RPE cells. (A) ARPE-19 cells were treated with 600 μM H2O2 and indicated concentrations of peptides at the same time. No oxidative stress and oxidative stress without peptides served as negative and positive controls, respectively. After 16 hours, cells were fixed with methanol and stained with Hoechst stain to visualize pyknotic nuclei. Representative images from each condition are shown. Scale bar: 100 μm. (B) Photos of five random fields per well were taken; the numbers of pyknotic and total number of cells were quantified using ImageJ software (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA), and the number of pyknotic cells was plotted as the percentage of total number of cells. (C) Plot of relative cell numbers determined using intracellular biomarkers–protease activity for dead cells (shown on left axis) and ATP for live cells (shown on right axis) as a function of P1 concentration is shown for ARPE-19 cells treated with indicated concentration of H2O2. Relative cell numbers are averages of duplicate wells ± SD (error bars). (D) Quantitation of relative dead cells as in (C) with increasing concentration of P1 and scrambled P1 (SCR) for ARPE-19 cells treated with 600 μM H2O2. Relative cell numbers of individual wells are shown in box-and-whiskers plots. The horizontal line inside the box is the median. (E) Bright-field photograph of primary pig RPE cells in culture. Scale bar: 100 μm. (F) Quantitation of live and dead cells as in (C) in primary pig RPE culture with indicated concentration of H2O2 and at indicated time. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005. Each experiment was performed more than two times.