Fig. 3.
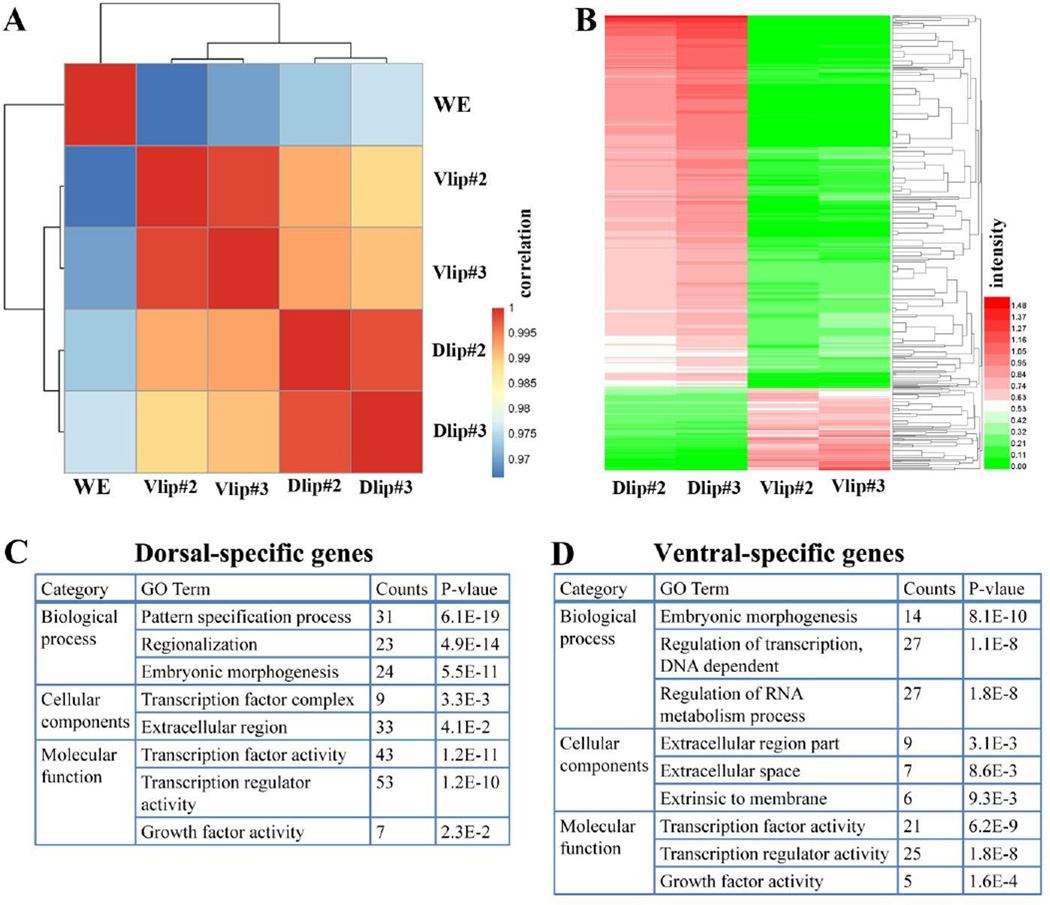
The D-V transcriptomes obtained by RNA-seq are highly reproducible. (A) Correlation plot of duplicate dorsal and ventral lip RNA-seq experiments from different egg clutches shows high reproducibility of the method. The correlation scores were calculated as the Pearson Correlation Coefficient (PCC) and color-coded as shown in the scale bar on the right of the panel. Note that Vlip and Dlip samples clustered into groups based on PCC. (B) Heatmap of differentially expressed genes between dorsal and ventral lip libraries. The expression data (RPKM) of each gene across different samples was scaled and represented as z-scores. The expression levels (expressed as logarithm of RPKM) are indicated in the scale bar on the right panel. (C) Table showing the top gene ontology (GO) terms associated with dorsal-specific genes. (D) Table of GO terms for ventral-specific transcripts.