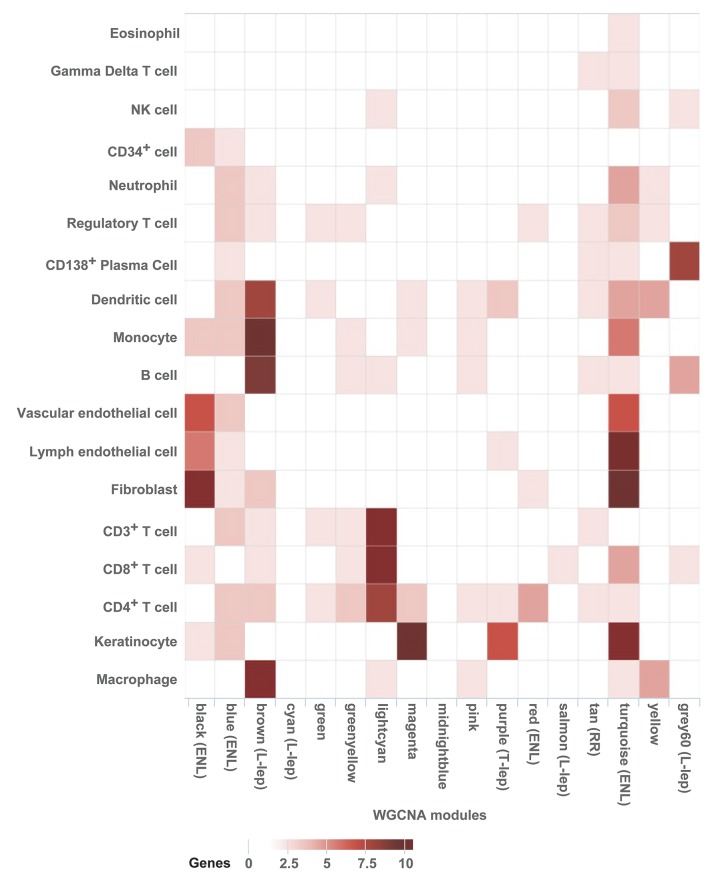
Figure 4. Integration of WGCNA gene modules with cell-type–specific gene signatures.
For each of the 17 modules of related genes derived from WGCNA analysis, enrichment for cell-type–specific gene signatures for 18 cell types with immune or structural functions were calculated and displayed in a heatmap of Z scores. Z scores were calculated from log2 fold change enrichment scores, using average gene expression for each leprosy subtype for each cell type. Cell-type names are provided in rows, and WGCNA module names from Figure 2 are provided in the column. Modules that were significantly associated with one leprosy subtype are labeled. ENL, erythema nodosum leprosum; L-lep, lepromatous leprosy; RR, reversal reaction; T-lep, tuberculoid leprosy.