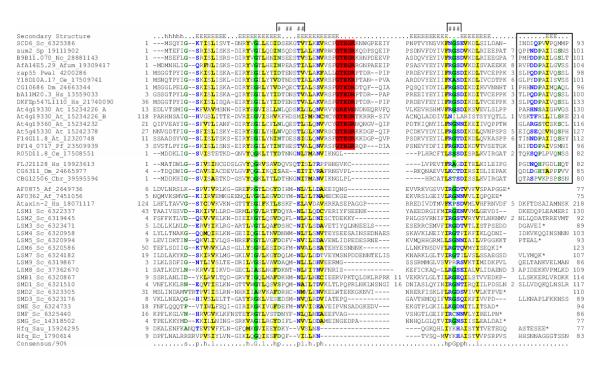
Figure 1.
Multiple alignment of the Scd6p family with representatives of other Sm domains. Multiple sequence alignment of the Sm domain of the Scd6p family was constructed using T-Coffee after parsing high-scoring pairs from PSI-BLAST search results. The secondary structure from the crystal structures is shown above the alignment with E representing a strand. The 90% consensus shown below the alignment was derived using the following amino acid classes: hydrophobic (h: ALICVMYFW, yellow shading) and its aliphatic subset (l: ALIV, yellow shading); small (s: ACDGNPSTV, green); and polar (p: CDEHKNQRST, blue). The limits of the domains are indicated by the residue positions, on each end of the sequence. A '*' denotes the end of the protein sequence. The numbers within the alignment are non-conserved inserts that have not been shown. The conserved GTEx+ motif of the scd6p family is shaded red. The residues involved in RNA binding are denoted by '#'s on the top of the aligment. The conserved C-terminal extension of the Scd6p family is shown in a box. The sequences are denoted by their gene name followed by the species abbreviation and GenBank Identifier (gi). The species abbreviations are: Af – Archaeoglobus fulgidus; Ec – Escherichia coli; Sau – Staphylococcus aureus; Afum – Aspergillus fumigatus; At – Arabidopsis thaliana; Cbr – Caenorhabditis briggsae; Ce – Caenorhabditis elegans; Dm – Drosophila melanogaster; Hs – Homo sapiens; Nc – Neurospora crassa; Pf-Plasmodium falciparum; Pwal – Pleurodeles waltl; Sc – Saccharomyces cerevisiae; and Sp – Schizosaccharomyces pombe.