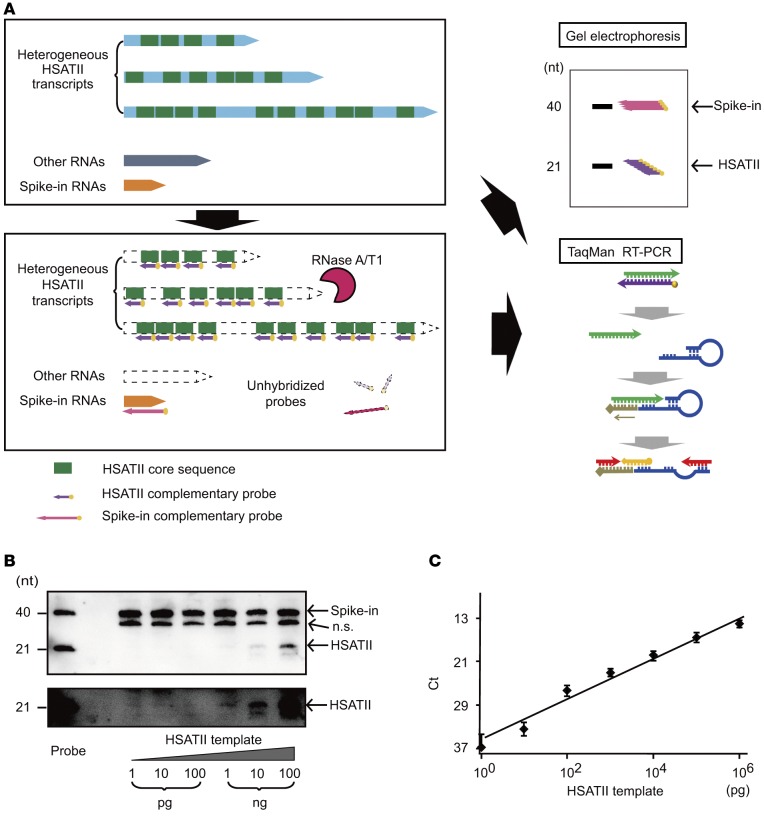
Figure 1. Tandem repeat amplification with nuclease protection method.
(A) Conceptual scheme of the tandem repeat amplification with nuclease protection (TRAP) method. Human satellite II (HSATII) RNAs are heterogeneous transcripts containing random repeats of the core sequences (upper panel). After hybridizing complementary probes to the core sequences, single-stranded RNAs including unhybridized probes are digested by nuclease A and T1 (lower panel). Protected core sequences can be accumulated and visualized by gel electrophoresis or quantified by TaqMan reverse transcription PCR (RT-PCR, right panels). (B) Detection of synthesized HSATII transcripts by gel electrophoresis after the TRAP method. Ten-fold serial dilutions from 1 pg to 100 ng of HSATII templates were examined. Spike-in probes were used as normalized controls and were detected at the size of 40 nucleotides (nt) and HSATII probes at the level of 21 nt. The lower panel shows high-contrast images of the bands of HSATII RNA in the upper panel. n.s., nonspecific bands. (C) A standard curve reflecting the accurate quantitation of the synthesized HSATII transcripts. Serial dilutions from 1 pg to 1 μg of HSATII RNA templates were quantified by TaqMan real-time PCR after performing the TRAP method. The standard curve is shown as a black line. Data represent the mean ± SD of 3 independent experiments.