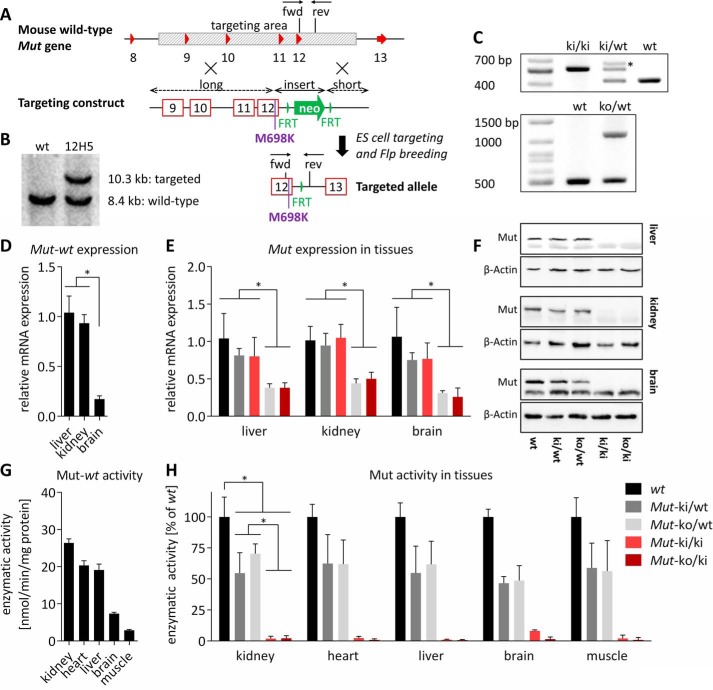
FIGURE 1.
Generation of KI allele and initial characterization. A, based on the wt mouse Mut gene, a targeting construct was generated to replace the Mut gene region spanning from introns 8 to 12 (shaded in gray), consisting of long and short arms of homology, as well as an insert with the missense mutation (p.Met698Lys) and a neomycin cassette (neo, green arrow) flanked by FRT sites (vertical green triangles). Homologous recombination and Flp deletor breeding resulted in the final targeted KI allele with the mutation and one remaining FRT site. Exons are represented by red triangles or boxes. Arrows marked fwd and rev indicate forward and reverse genotyping primers, respectively. B, Southern blotting of targeted clone 12H5. C, upper panel, genotyping PCR of ear biopsy DNA for the KI allele with forward and reverse primers depicted in A. The asterisk indicates an additional heterodimer band as confirmed by Sanger sequencing. Lower panel, genotyping PCR of KO allele. D and E, Mut expression among wt tissues (error bars are S.E.; n = 4; *, p < 0.01) (D) and KO allele-dependent loss of Mut expression (bars are means normalized to wt in each tissue; error bars depict S.E.; n ≥ 4; *, p < 0.05) (E). F, Western blot analysis of Mut protein levels using β-actin as loading control. The lower band in the top panel of each organ represents an unspecific band. G and H, Mut activity varies by tissue (G) and by genotype (H) (bars are mean values and in each tissue normalized to the wt value; error bars are S.E.; n ≥ 4; *, p < 0.0001). Significance levels are the same in all five tissues (not depicted for clarity).