Abstract
Objective
To develop and validate a model of incident type 2 diabetes based solely on administrative data.
Data Sources/Study Setting
Optum Labs Data Warehouse (OLDW), a national commercial administrative dataset.
Study Design
HealthImpact model was developed and internally validated using nested case–control study design; n = 473,049 in training cohort and n = 303,025 in internal validation cohort. HealthImpact was externally validated in 2,000,000 adults followed prospectively for 3 years. Only adults ≥18 years were included.
Data Collection/Extraction Methods
Patients with incident diabetes were identified using HEDIS rules. Control subjects were sampled from patients without diabetes. Medical and pharmacy claims data collected over 3 years prior to index date were used to build the model variables.
Principal Findings
HealthImpact, scored 0–100, has 48 variables with c‐statistic 0.80815. We identified HealthImpact threshold of 90 as identifying patients at high risk of incident diabetes. HealthImpact had excellent discrimination in external validation cohort (c‐statistic 0.8171). The sensitivity, specificity, positive predictive value, and negative predictive value of HealthImpact >90 for new diagnosis of diabetes within 3 years were 32.35, 94.92, 22.25, and 96.90 percent, respectively.
Conclusions
HealthImpact is an efficient and effective method of risk stratification for incident diabetes that is not predicated on patient‐provided information or laboratory tests.
Keywords: Diabetes mellitus type 2, risk assessment/methods, theoretical models, decision support techniques
In the United States, more than 1.5 million adults are newly diagnosed with diabetes each year (CDC 2012). An additional 79 million adults, or 37 percent of U.S. adults, are estimated to be at risk for developing diabetes (CDC 2011), historically defined by fasting blood glucose values just below the diagnostic threshold for diabetes (e.g., prediabetes) (Tamez‐Perez, Proskauer‐Pena, and Hernandez‐Coria 2013; ADA 2015). Randomized controlled trials demonstrated that lifestyle modifications and pharmacotherapies can be effective in delaying or reversing the progression of hyperglycemia, reducing the personal and societal burdens of prediabetes and diabetes (Pan et al. 1997; Tuomilehto et al. 2001; Chiasson et al. 2002; Knowler et al. 2002, 2009; Nichols and Brown 2005; Herman et al. 2012). A major barrier to implementation of diabetes risk reduction programs is the difficulty of identifying individuals at risk. Many public health efforts to screen patients for diabetes have been hindered by low turnout, high cost incurred by testing a large number of people, and lack of opportunities to provide counseling or referral (Engelgau, Narayan, and Herman 2000; Tabaei et al. 2003). As a result, only 11 percent of people with prediabetes are estimated to be aware of their condition (CDC 2013).
Current clinical practice guidelines recommend using laboratory criteria to diagnose prediabetes: fasting glucose (FPG) 100–125 mg/dL, 2‐hour glucose 140–199 mg/dL after an oral glucose tolerance test (OGTT), or glycosylated hemoglobin (HbA1c) 5.7–6.4 percent (ADA 2015). In an effort to better identify high‐risk individuals, more than 145 risk models for type 2 diabetes have been proposed over the past decade (Noble et al. 2011; Abbasi et al. 2012; Kengne et al. 2014), but they have not been incorporated into routine clinical practice in part due to the prohibitive effort of obtaining necessary clinical, laboratory, and/or patient‐provided information such as anthropometric measurements, lifestyle information, smoking, family history, education, income, and/or laboratory data (Noble et al. 2011; Abbasi et al. 2012; Kengne et al. 2014). This information cannot be obtained without direct patient contact, manual data processing, or invasive and resource‐intensive laboratory tests, making currently available models inadequate to fully meet broader public health needs.
Our goal was to develop and both internally and externally validate an efficient risk prediction model for incident type 2 diabetes (HealthImpact) that is based entirely on administrative data, thereby reducing reliance on direct patient contact and obviating need for patient‐provided information and laboratory tests. Importantly, our objective was not to identify patients with undiagnosed diabetes or to estimate the probability that a specific individual will develop diabetes, but rather to identify people whose risk of diabetes is sufficiently high to warrant further evaluation (Box S1). Such a model may be used by health systems or accountable care organizations (ACOs) as part of population health management efforts to identify and reach out to at‐risk patients or to facilitate resource allocation based on anticipated need; it may also be used by payers seeking to facilitate risk‐adjustment across health plans. In addition, as primary care groups are held increasingly responsible for population health management, a tool like HealthImpact can be useful, though it may need to be operated and maintained by a contracted external entity.
While a risk model that utilizes only claims data would be limited to insured individuals with at least a minimal interaction with the health care system, 79 percent of American adults between ages 19–64 years now report having health insurance coverage and more are expected to gain coverage as a result of the Affordable Care Act (Collins et al. 2013). Moreover, because HealthImpact relies on the near‐universally used International Classification of Diseases (ICD) diagnostic codes and National Drug Codes (NDC), its algorithm is generalizable to any insurance type and health system, including other private payers, public payers (e.g., Medicare, Medicaid, Veterans Administration), and international health systems. Similarly, ongoing efforts to standardize and extend Health Information Exchanges (HIEs) in accordance with Meaningful Use (2013a), and the upcoming implementation of the ICD‐10 format, could also be leveraged for HealthImpact adaptation for future use.
Methods
Dataset
This study utilizes data between 1994 and 2012 from the Optum Labs Data Warehouse (OLDW) (Wallace et al. 2014), a national deidentified dataset of more than 100 million privately insured individuals that is geographically and racially diverse, including individuals of all ages (including Medicare Advantage beneficiaries ≥65 years old) and from all 50 states, with greatest representation in the Midwest and South U.S. Census Regions (2014). OLDW provides full access to professional, facility, and outpatient prescription medication claims. Patient‐identifying information was encrypted or removed from the study database prior to its release to the study investigators, such that it is compliant with HIPAA and exempt from Institutional Review Board review.
Definition of Outcome
The study outcome was a binary variable indicating a new diagnosis of diabetes mellitus (e.g., incident diabetes on the next day); diabetes was defined by Healthcare Effectiveness Data and Information Set (HEDIS) diagnosis and medications criteria (NCQA 2009). We required that individuals meet at least two HEDIS criteria within 180 days to exclude diabetes rule‐out cases, but diagnosis date was the date of the first HEDIS‐qualifying claim. Finally, to ensure that identified individuals have incident rather than prevalent diabetes, we required a 36‐month period of continuous enrollment without diabetes‐related claims or medications before the first HEDIS‐qualifying event.
Development and Internal Validation
The HealthImpact model was developed and internally validated using nested case–control study design, utilizing stratified random sampling as described in Figure 1 and online‐only Supplement. This enriched the target population for diabetes cases and thereby optimized model development (Box S2).
Figure 1.
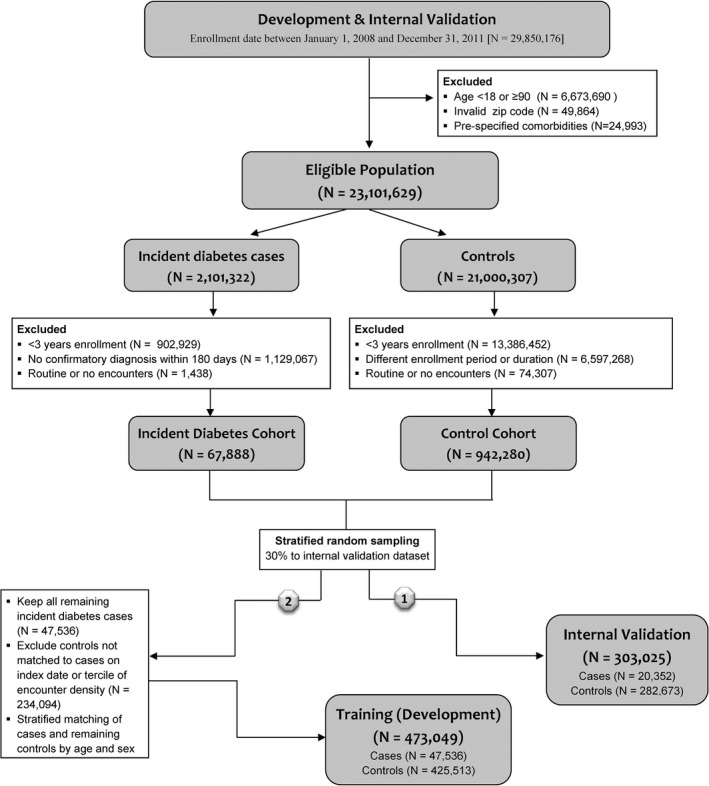
Study Design. HealthImpact Development and Internal Validation
First, we identified adults, aged 18–89 years, with ≥36 months of continuous enrollment, and ≥3 claims within a qualifying health plan between January 1, 2008, and November 30, 2012. We performed complete‐case analysis, including only individuals with uninterrupted enrollment for ≥36 months. A qualifying health plan was defined as a plan having ≥1,000 total medical and ≥1,000 total drug claims, and claims available during ≥9 distinct months over a 3‐year period. The rationale for excluding health plans with insufficient medical and/or drug claims was to exclude outlier health plans that are not representative of the health insurance marketplace.
Step‐wise demonstration of how the study population was assembled is depicted in Figure 1. There were 29,850,176 individuals with ≥36 months of uninterrupted enrollment between January 1, 2008, and November 30, 2012. We excluded 6,673,690 individuals aged <18 or ≥90 years; 49,864 individuals with invalid zip codes (recorded zip code did not match list of possible zip codes available from the U.S. Postal Service); and 24,993 individuals with diagnoses of secondary diabetes (ICD‐9 codes 249.x), disorders of pancreatic internal secretion (ICD‐9 code 251.8), and poisoning by adrenal cortical steroids (ICD‐9 code 962.0) during the 36‐month enrollment period. This resulted in 23,101,629 eligible individuals, of whom 2,101,322 had a diagnosis of diabetes (“incident diabetes cases”) during the 36‐month enrollment period.
In the incident diabetes cohort, we excluded 1,129,067 people who did not have a confirmatory diagnosis of diabetes within 180 days of the first diagnosis code or medication claim meeting HEDIS criteria (NCQA 2009) to exclude diabetes rule‐out cases. To ensure identification of incident rather than prevalent diabetes cases, we then excluded 1,129,077 individuals who had one or more diabetes diagnosis claims meeting HEDIS criteria during ≥3 years prior to the date of first diabetes diagnosis. Finally, to minimize bias from inclusion of individuals who had no encounters with the health care system, we excluded 1,438 people with no claims or with routine care only (ICD‐9 codes v20 [well‐child visit], v25 [contraception], v70‐82 [general health and screenings]) (King and Zeng 2001). Ultimately, there were 67,888 people in the incident diabetes cohort. Index date for the cases was the date of the first HEDIS‐qualifying claim, and all cases had at least 36 months of observation prior to that date. However, variables from only the 36 months immediately preceding the index date were considered in all analyses.
We applied the same inclusion/exclusion criteria to eligible individuals without diabetes (“controls”), resulting in 942,280 people. The control cohort was designed to match the case cohort on the distributions of enrollment duration, as described in Box S3, to minimize risk of sampling and selection biases. Controls were matched without replacement. For the controls, as for the cases, only variables from the 36 months immediately preceding the index date were considered in the analyses.
Drawing from the incident diabetes and control cohorts, we first assembled the internal validation population by stratified random sampling to allocate 30 percent of the incident diabetes and control cohorts to the internal validation dataset; this yielded a population of 20,352 cases and 282,673 controls (total n = 303,025). We stratified the internal validation cohort by sex and age group (5‐year increments from 25–29 to 75–79 years, and wider end groups for 18–24 and 80–89 years). In the internal validation population, the cases and controls were matched solely on enrollment duration.
All of the remaining 47,536 people within the incident diabetes cohort were assigned to the HealthImpact training (development) population. The remaining 559,607 controls were stratified matched to these 47,536 cases on index year (2008, 2009, 2010, 2011) and tercile of encounter density (<20, 20–48, 48–86, ≥86) as described in Box S3; 282,673 could not be matched and were excluded. Each case with incident diabetes could have up to 10 matched controls, matched not only on enrollment duration (as in the internal validation dataset) but also on encounter density and index year. Encounter density was measured as the fraction of unique dates with a claim, and it was included to minimize bias from diagnostic access. Additional matching on index year accounted for potential changes in secular trends and clinical practices. The training dataset included 473,049 people (47,536 cases and 425,513 controls).
External Validation
We identified a random cohort of 2,000,000 adults (18–79 years) from a population of 7,908,918 people in the OLDW enrolled on April 1, 2009, who had continuous enrollment for ≥12 months (April 1, 2008, to April 1, 2009) and had no documentation of diabetes mellitus, secondary diabetes, disorders of pancreatic internal secretion, or poisoning by adrenal cortical steroids during that ≥12‐month period. All individuals were followed prospectively until disenrollment or March 30, 2012, whichever came first. To maximize generalizability of HealthImpact, there were no exclusions based on type of health plan, or number and type of encounters. HealthImpact scores were calculated using data available as of April 1, 2009. Patients were followed prospectively, and point prevalence of diabetes was calculated at 12, 24, and 36 months of continuous enrollment, with the denominator being all people with uninterrupted enrollment to that date. Individuals were censored upon disenrollment from the health plan, defined as the absence of medical or pharmacy coverage for ≥31 consecutive days. Censored patients were included prior to the time of censure, such that there was no missing information at the time of analysis.
Comparison of HealthImpact to Laboratory Methods: A Sensitivity Analysis
There were 426,746 individuals in the external validation cohort who had available laboratory data; laboratory data availability within OLDW is contingent on data sharing agreements between Optum Labs and clinical laboratories. Of them, 15,595 were excluded because they had laboratory evidence of diabetes at baseline, defined by FPG ≥126 mg/dL, 2‐hour glucose after an OGTT ≥200 mg/dL, or HbA1c ≥6.5 percent (ADA 2015). Individuals with prediabetes were identified on the basis of FPG 100–125 mg/dL, 2‐hour glucose after OGTT 140–199 mg/dL, or HbA1c 5.7–6.4 percent (ADA 2015). The sensitivity, specificity, PPV, and NPV of each incident diabetes prediction method were calculated using point prevalence of diabetes at 1, 2, and 3 years among those individuals who had no diabetes at baseline.
Independent Variables
For HealthImpact development and internal validation, we collected claim and enrollment data for matched individuals from 3 years prior to the date of cohort entry; and for external validation, for 1 year prior to cohort entry. These included diagnoses (ICD‐9 codes), generic drug names (NDC codes), zip code of residence, age, and sex (Box S4). Race/ethnicity and income were derived by proxy by linking residential addresses to a U.S. Census zip code tabulation area (ZCTA) group (Krieger et al. 2003). Each ZCTA group was assigned a race tercile for lowest, middle, and highest percentage of non‐white population with tercile cut‐offs of 9 and 24 percent non‐white population to reflect the higher prevalence of diabetes in the minority population. Age was modeled as bands of 5‐year increments from 25–29 to 75–79 years, and wider end groups for 18–24 and 80–89 years. These parameters were used to maximize generalizability of the HealthImpact model, taking into consideration different capabilities and compositions of other administrative datasets.
Logical Observation Identifiers Names and Codes (LOINC) codes were used to identify laboratory study results in the external validation subpopulation: OGTT (1504‐0, 1518‐0, 20437‐0), fasting glucose (1558‐6), random glucose (2339‐0, 32016‐8, 41653‐7, 2345‐7), and HbA1c (4548‐4, 17856‐6).
Analytic Methods
There were 12,482 clinical, pharmaceutical, and enrollment variables that could be included in the HealthImpact model (Box S4). Because many of these variables were likely to be correlated, we first grouped them using the Agglomerative Single‐Link Clustering Algorithm (Jain, Murty, and Flynn 1999), which uses a series of decreasing correlation thresholds to determine all sets of predictors that have pair‐wise correlations greater than a prespecified threshold of 0.70. After clustering 113 variables into 34 groups (Table 1), remaining 12,405 variables were passed to GLMNET to solve with a pure lasso logistic regression model as described in Box S5 (Friedman, Hastie, and Tibshirani 2010). The distribution of the number of available nondemographic variables for each person in the training and internal validation cohorts is shown in Figure S1; 90 percent had ≥4 variables and 10 percent had >52 variables.
Table 1.
HealthImpact Model: Demographic, Clinical Diagnosis, Pharmacy, and Clinical Group Variables Included in the Final HealthImpact Model
| Type | Variable Description | Coefficient |
|---|---|---|
| Constant | Baseline (female, 35–39 years, medium minority) | −1.251 |
| demographic | Age: 18–24 | −1.323 |
| Age: 25–29 | −0.592 | |
| Age: 30–34 | −0.231 | |
| Age: 40–44 | 0.130 | |
| Age: 45–49 | 0.311 | |
| Age: 50–54 | 0.494 | |
| Age: 55–59 | 0.571 | |
| Age: 60–64 | 0.639 | |
| Age: 65–69 | 0.686 | |
| Age: 70–74 | 0.853 | |
| Age: 75–79 | 0.809 | |
| Age: 80–89 | 0.617 | |
| Gender: male | 0.317 | |
| Low minority | −0.154 | |
| High minority | 0.454 | |
| ICD‐9‐CM | Intestinal disaccharidase deficiencies and disaccharide malabsorption (271.3) | 1.247 |
| Dysmetabolic syndrome X (277.7) | 1.415 | |
| Obstructive sleep apnea (327.23) | 0.135 | |
| Benign hypertensive heart disease without heart failure (402.10) | 0.417 | |
| Coronary atherosclerosis of native coronary artery (414.01) | 0.120 | |
| Congestive heart failure, unspecified (428.0) | 0.262 | |
| Acute respiratory failure (518.81) | 0.809 | |
| Other chronic nonalcoholic liver disease (571.8) | 0.633 | |
| Other acne (706.1) | −0.244 | |
| Hypersomnia with sleep apnea, unspecified (780.53) | 0.210 | |
| Unspecified sleep apnea (780.57) | 0.187 | |
| Polydipsia (783.5) | 1.742 | |
| Shortness of breath (786.05) | 0.154 | |
| Other dyspnea and respiratory abnormalities (786.09) | 0.115 | |
| Other abnormal blood chemistry (790.6) | 0.983 | |
| Polycystic ovaries (256.4) | 1.599 | |
| Glycosuria (791.5) | 2.464 | |
| Diphtheria–tetanus–pertussis, combined (v06.1) | −0.388 | |
| Medication | Amlodipine besylate | 0.165 |
| Furosemide | 0.439 | |
| Teriparatide | 2.790 | |
| Merged groups | Benign neoplasm of skin (216.5, 216.9) | −0.434 |
| Delivery (650, v270) | −1.224 | |
| Abnormal glucose (790.2, 790.29) | 1.992 | |
| Ethinyl estradiol (multiple agents) | −0.310 | |
| Fenofibrate (multiple agents) | 0.532 | |
| Abnormal maternal glucose tolerance (648.8) | 3.671 | |
| Hyperlipidemia (272.2, 272.4) | −0.023 | |
| Hypertension (401.0, 40.1.1, 401.9) | 0.594 | |
| Nonallopathic lesion (739.0–739.4) | −0.422 | |
| Overweight/obesity (278.0, 278.00–278.02) | 0.760 | |
| Impaired glucose (790.21, 790.22) | 1.659 |
Note. In parentheses are shown the specific ICD‐9 codes used to identify these variables.
Model analysis and accuracy measures used the caret, GLMNET and RMS packages from the R statistical library, v3.1.1 (2013b; Harrell 2014). GLMNET implements lasso and regularized regression methods that allow for variable selection in large datasets. GLMNET's optimization path allows it to find the best fit for each unique number of variables, 1 to N. Specifically, due to concern for overfitting of the data when working with very large populations and a large number of variables, variable selection was performed with eight‐way cross‐validation (CV) so that each fit with n variables was evaluated against seven overlapping datasets. For each fit, the c‐statistic is measured on the held out CV‐sample (Friedman, Hastie, and Tibshirani 2010). Details of model fitting are discussed in the Online‐only Supplement.
Internal validity of the model was established through both cross‐validation during variable selection and analysis of the model fit using bootstrapping. The 47 variable model had a mean AUC (area under the receiver operating characteristic curve) of 79.43 within [79.3, 79.5] for eight‐fold cross‐validation, and 80.8 percent when refit to the entire training population. The training data fit was also evaluated using the RMS package validate function, which performed 100 bootstrap fits of the data to compute bias‐corrected estimates of the c‐statistic. The original c‐statistic was 0.8082, and the bias‐corrected c‐statistic was 0.80815. The Brier statistic, the average mean square error between the predicted and actual values for training data, was 0.1748 (bias‐corrected, 0.1747). The training data R 2 value was 0.3604 (bias‐corrected, 0.3600). The Yates slope, the difference in mean predicted values for the incident and control populations, for the training data was 0.2780; this statistic is not part of the model validation suite of measures and therefore is not bias‐corrected. Finally, each variable was tested for colinearity (Harrell 2001), and the final model had maximum variance inflation factor 2.59.
Because the training and internal validation sets were built retrospectively without the ability to compute a true population prevalence, we did not compute the calibration of the fits. This calibration would show a fit to a prevalence in the data that cannot reflect a true prevalence. In place of this calibration, the external dataset was validated by examining 1, 2, and 3 year incidence of diabetes as predicted by the HealthImpact score.
Results
HealthImpact model Development and Internal Validation
There were 473,049 individuals in the HealthImpact training dataset (47,536 cases and 425,513 controls) and 303,025 individuals in the internal validation dataset (20,352 cases and 282,673 controls); see Figure 1. Baseline clinical and demographic characteristics of the training and internal validation study populations are summarized in Table 2. The training population was, on average, 45.76 years old (SD, 13.65), 44.74 percent male, and with a relatively high prevalence of hypertension (29.66 percent) and hyperlipidemia (36.38 percent). The internal validation population was slightly younger at 44.21 years (SD, 13.67) and had lower prevalence of both hypertension (23.84 percent) and hyperlipidemia (30.84 percent); 47.90 percent were male. The relative proportion of patients from high minority zip codes was comparable in the training (35.54 percent) and internal validation (36.34 percent) cohorts.
Table 2.
Study Cohorts: Baseline Characteristics of People Included in the HealthImpact Training (Development), Internal Validation, and External Validation Cohorts
| Training (N = 473,049) | Internal Validation (N = 303,025) | External Validation (N = 2,000,000) | |
|---|---|---|---|
| Age, years, mean (SD) | 45.76 (13.65) | 44.21 (13.67) | 44.12 (12.66) |
| Gender, male, N (%) | 211,638 (44.74) | 145,148 (47.90) | 967,243 (48.36) |
| Enrollment, years, mean (SD) | |||
| Prior to diagnosis | 5.56 (2.06) | 5.51 (2.03) | 3.65 (2.56) |
| Following diagnosis | 1.84 (1.20) | 1.83 (1.22) | 2.11 (1.26) |
| Race/ethnicity, N (%) | |||
| High (>24%) minority | 168,124 (35.54) | 110,110 (36.34) | 761,026 (38.05) |
| Medium (9–24%) minority | 175,568 (37.11) | 111,461 (36.78) | 718,080 (35.90) |
| Low (<9%) minority | 168,124 (35.54) | 110,110 (36.34) | 520,894 (26.04) |
| Comorbidities, N (%) | |||
| Ischemic heart disease | 29,678 (6.27) | 14,343 (4.73) | 85,142 (4.26) |
| Cerebrovascular disease | 16,189 (3.42) | 7,967 (2.63) | 43,496 (2.17) |
| Peripheral vascular disease | 10,225 (2.16) | 4,921 (1.62) | 27,334 (1.37) |
| Hypertension | 140,297 (29.66) | 72,234 (23.84) | 427,801 (21.39) |
| Hyperlipidemia | 172,103 (36.38) | 93,438 (30.84) | 563,822 (28.19) |
| Obesity | 34,036 (7.20) | 18,238 (6.02) | 92,694 (4.63) |
| Elevated blood glucose | 16,441 (3.48) | 7,883 (2.60) | 28,572 (1.43) |
| Gestational diabetes | 3,522 (0.74) | 1,847 (0.61) | 10,001 (0.50) |
| Polycystic ovarian syndrome | 3,413 (0.72) | 1,761 (0.58) | 10,750 (0.54) |
| Medication number, mean (SD) | 1.32 (1.83) | 1.00 (1.62) | 0.85 (1.49) |
| Clinical encounters per year, mean (SD) | |||
| Prior to diagnosis | 4.88 (6.16) | 3.97 (5.31) | 3.72 (4.98) |
| Following diagnosis | 10.26 (14.27) | 8.58 (12.47) | 9.50 (13.19) |
| Observation duration, months, median (IQR) | 62.8 (47.6, 86.8) | 62.8 (47.6, 86.8) | 38.3 (21.1, 53.6) |
Characteristics of the incident diabetes and control cohorts prior to stratified random allocation to the training and internal validation datasets are shown in Table S1. Patients in the control group were selected to match patients in the incident diabetes group only on their enrollment duration, while other characteristics were permitted to vary to make all potentially predictive characteristics eligible for consideration of inclusion in the HealthImpact model. Patients in the incident diabetes cohort were older (52.35 years vs. 44.47 years) with higher proportion of men (52.40 percent vs. 45.36 percent) and patients from high minority zip codes (44.50 percent vs. 35.02 percent). They also had greater prevalence of comorbid metabolic and atherosclerotic diseases.
The HealthImpact model included 48 terms (a constant and 47 variables; Table 1), and it had bias‐corrected c‐statistic 0.80815 (Figure S2). We proposed three HealthImpact score thresholds to signify high, medium, and low risk of incident diabetes based on achieving sensitivity and specificity approaching 80 percent: 50, 75, and 90 (Table 3). Complete listing of sensitivities and specificities at each score threshold in the training dataset is available in Table S2. Clinical vignettes describing individuals with varying HealthImpact scores are included in Box S6.
Table 3.
HealthImpact Accuracy in the Training, Internal Validation, and External Validation Populations. HealthImpact Thresholds Were Chosen to Signify Low (50), Intermediate (75), and High (90) Risk of Incident Diabetes at Three Years
| HealthImpact Threshold | >50 | >75 | >90 |
|---|---|---|---|
| Training dataset (n = 473,049) | |||
| Sample size | 132,819 | 41,782 | 15,213 |
| Sensitivity | 69.23 | 38.10 | 17.86 |
| Specificity | 76.49 | 94.42 | 98.42 |
| Internal validation dataset (n = 303,025) | |||
| Sample size | 69,945 | 19,813 | 7,042 |
| Sensitivity | 69.18 | 37.74 | 18.01 |
| Specificity | 80.11 | 95.68 | 98.80 |
| External validation datasets | |||
| Year 1 (n = 1,383,743) | |||
| Sample size | 424,294 | 167,124 | 71,309 |
| Sensitivity | 80.54 | 53.70 | 31.07 |
| Specificity | 69.91 | 88.40 | 95.15 |
| Positive predictive value | 2.99 | 5.05 | 6.85 |
| Negative predictive value | 99.68 | 99.40 | 99.17 |
| Year 2 (n = 1,054,142) | |||
| Sample size | 342,925 | 139,977 | 61,203 |
| Sensitivity | 80.90 | 54.45 | 32.13 |
| Specificity | 68.73 | 87.79 | 94.88 |
| Positive predictive value | 6.30 | 10.40 | 14.03 |
| Negative predictive value | 99.28 | 98.67 | 98.17 |
| Year 3 (n = 827,969) | |||
| Sample size | 278,043 | 116,235 | 51,758 |
| Sensitivity | 80.88 | 54.52 | 32.35 |
| Specificity | 68.54 | 87.78 | 94.92 |
| Positive predictive value | 10.35 | 16.70 | 22.25 |
| Negative predictive value | 98.76 | 97.73 | 96.90 |
We internally validated HealthImpact among 303,025 individuals (Table 2). In this population, the c‐statistic was 0.8270 (Figure S2). The sensitivity of HealthImpact in the internal validation cohort was comparable to that in the training cohort: 69.18, 37.74, and 18.01 percent for HealthImpact >50, >75, and >90, respectively. The specificity was slightly better, particularly at lower HealthImpact scores: 80.11, 95.68, and 98.80 percent for HealthImpact >50, >75, and >90, respectively. The higher rate of false positives (e.g., lower specificity) in the training population is likely due to matching of cases and controls on their encounter density, which enriched the control group for individuals with comparable number of claims as people with incident diabetes. This makes it more difficult for the model to differentiate between the incident diabetes cases and the controls than in the unmatched internal validation population, where the controls tend to have fewer clinical encounters (Table 2). This is discussed further in Box S8.
HealthImpact External Validation
The HealthImpact model was externally validated in a population of 2,000,000 adults with no diagnosis of diabetes (Table 2). At the time of cohort entry, mean age was 44.12 years (SD, 12.66) and 48.36 percent were male. They were healthier than the training and internal validation populations with fewer comorbidities, medications, and clinical encounters. This was expected, as the HealthImpact training/internal validation population was enriched for people with higher health care utilization by requiring ≥3 years of enrollment with ≥1 nonroutine care clinical encounter. Moreover, the training/internal validation dataset was assembled to have 10 percent incidence of type 2 diabetes, but no such restriction was placed on the external validation dataset that was designed to mimic the general commercially insured population.
Distribution of HealthImpact scores at cohort entry and the corresponding 3‐year incidence rates of type 2 diabetes are shown in Figure 2. HealthImpact had good discrimination for incident diabetes at 1, 2, and 3 years with c‐statistic 0.8200, 0.8171, and 0.8171, respectively. The Brier scores at the same intervals are 0.177, 0.179, and 0.180; Yates slopes are 0.3163, 0.3130, and 0.3122, respectively. The higher c‐statistics in the external validation dataset compared to the training and internal validation datasets are not unexpected (Steyerberg 2009), and they may be due to more noncontributory data (“noise”) in the latter datasets that had longer periods of preceding enrollment. The sensitivity of HealthImpact for predicting incident diabetes at 1, 2, and 3 years was also markedly higher in the external validation dataset, ranging 80.54–80.90 percent for HealthImpact >50; 53.70–54.52 percent for HealthImpact >75; and 31.07–32.35 percent for HealthImpact >90. Specificity was mildly decreased in the external validation dataset, ranging 68.54–69.91 percent for HealthImpact >50; 87.78–88.40 percent for HealthImpact >75; and 94.92–95.15 percent for HealthImpact >90. The PPV approached 10.35 percent at year 3 for HealthImpact >50, 16.70 percent for HealthImpact >75, and 22.25 percent for HealthImpact >90. NPV was 97 percent or higher at all score thresholds and time periods (Table 3).
Figure 2.
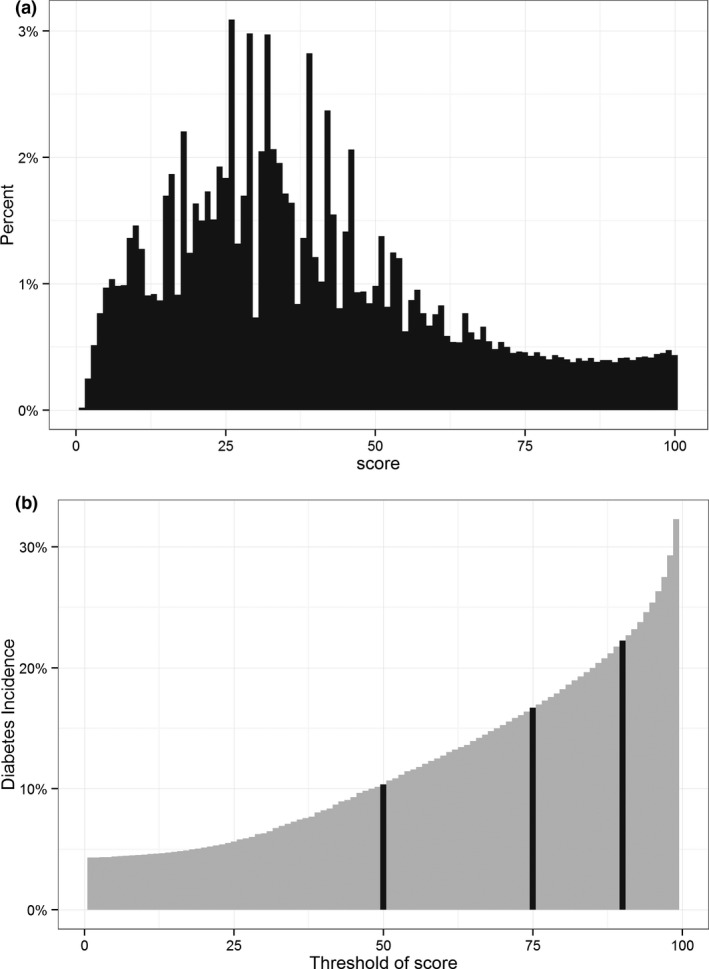
(a) Distribution of Baseline HealthImpact Scores in the External Validation Dataset among People without Diagnosed Diabetes (n = 2,000,000). (b) Cumulative Incidence of Diabetes at 3 years as a Function of Baseline HealthImpact Score.
Note. Black lines correspond to HealthImpact scores 50, 75, and 90. The cumulative incidence of diabetes at 3 years was 1.24 percent with HealthImpact score ≤50, 5.80 percent with HealthImpact score 50–75, 12.24 percent with HealthImpact score 75–90, and 22.25 percent with HealthImpact score >90.
We also examined the time to diabetes diagnosis based on baseline HealthImpact score, as sufficient lead time to diagnosis is important to ensure timeliness of patient identification and potential for disease‐modifying intervention. Details and results of this analysis are presented in Box S7, Table S3, and Figure S3. Among patients with HealthImpact score 90–100, 6.1, 11.7, and 17.1 percent developed diabetes by 1, 2, and 3 years, respectively. Among patients with baseline HealthImpact 75–90, the cumulative incidence of diabetes was 3.2, 5.9, and 8.7 percent by 1, 2, and 3 years, respectively. Among patients with baseline HealthImpact 50–75, the cumulative incidence of diabetes was 1.3, 2.6, and 3.8 percent by 1, 2, and 3 years, respectively. Almost no patients (<1 percent) with baseline HealthImpact score <50 developed diabetes by 3 years.
We conducted a sensitivity analysis comparing HealthImpact to laboratory measurements of glycemic control (e.g., fasting glucose, HbA1c, and OGTT), described in detail in Box S9. There were 421,520 people among the 2,000,000 people included in the external validation dataset who had laboratory studies performed and available at baseline (Table S4). Comparison of sensitivities, specificities, PPVs, and NPVs of HealthImpact and the laboratory definition of prediabetes are presented in Table S4. Overall, HealthImpact had lower sensitivity and higher specificity than laboratory testing at higher thresholds (HealthImpact >75), with similar NPV and better PPV.
Discussion
The increasing burden of diabetes and diabetes‐related complications on individuals, society, and the health care system has spurred multifaceted prevention programs aimed at people at highest risk for developing diabetes. These efforts have been hindered by the challenges of identifying high‐risk patients in a reliable and cost‐effective manner that could be implemented on both the clinic and population levels. Current clinical practice guidelines use laboratory criteria of prediabetes to identify individuals at increased risk for developing diabetes (Tamez‐Perez, Proskauer‐Pena, and Hernandez‐Coria 2013; ADA 2015), but these can be time‐consuming, inconvenient, costly, or altered by factors other than glycemia (Sacks 2011). Noninvasive diabetes risk prediction models have required clinical and patient‐provided information that necessitate patient contact or electronic documentation that exceeds the analytic capabilities of many current EMR systems (Noble et al. 2011). We therefore developed and validated a novel diabetes risk prediction algorithm based entirely on administrative data, which may be implemented by any health system that uses billing data or electronic health records.
HealthImpact has good discrimination for incident diabetes, with c‐statistic >0.8 when validated in the general population of commercially insured adults in the United States. HealthImpact therefore performed better than, or comparable to, previously published invasive and noninvasive diabetes prediction models (Abbasi et al. 2012; Kengne et al. 2014). However, in contrast to previously available models, HealthImpact is not predicated on information that could only be obtained through direct patient contact such as family history, anthropomorphic measurements (BMI, height, weight, waist circumference), or risk factor information (smoking status, dietary habits, patterns of physical activity). The HealthImpact PPV with just 3 years of follow‐up is higher than that of alternative models with 5 to 10 years of follow‐up, despite lacking patient‐provided or anthropomorphic variables that were included to improve the other models. HealthImpact has 3‐year PPVs 10, 16, and 22 percent at score thresholds 50, 75, and 90, respectively. The Finish Diabetes Risk score had a 5‐year PPV of 10 percent at the proposed threshold value of 9, which has comparable sensitivity and specificity to HealthImpact threshold value of 90 (Lindström and Tuomilehto 2003). The Australian AUSDRISK score had a 5‐year PPV of 13 percent (Chen et al. 2010). The Canadian DPoRT score had a 5‐year PPV 4 percent for men and 3 percent for women (Balkau et al. 2008). The German Diabetes Risk score had 5‐year PPV between 6 and 11 percent for their proposed score thresholds (Schulze et al. 2007). Moreover, while previously published models have predicted risk of incident diabetes at 5–10 years of follow‐up, HealthImpact can be used to identify patients at more proximal risk—as short as 1 year—and therefore be more timely and relevant to ongoing clinical practice, payer risk management, and patient behavior. Its ability to predict risk with sufficient lead time to potentially intervene also makes it clinically relevant.
The OLDW dataset is unique due to its size, representation of a large segment of the U.S. population spanning all ages and demographics, and inclusion of multiple payers and health systems (Wallace et al. 2014). The observed rates of incident diabetes in risk strata predicted by HealthImpact are comparable to those using laboratory‐based criteria (Zhang et al. 2010; Ackermann et al. 2011). An important limitation is the fact that HealthImpact was developed and first validated among commercially insured adults in the United States, but not the general U.S. population, as OLDW does not include individuals insured by government payers (Medicare, Medicaid, VA, or Indian Health Service) or the uninsured. Nonetheless, while it may not be immediately generalizable to populations with different demographic and socioeconomic compositions, HealthImpact may be adapted to different environments because the requisite demographic, socioeconomic, geographic, and ethnic/racial information is incorporated into the model. Diabetes risk also varies throughout the world, and HealthImpact is one of few models developed and validated in the United States (Noble et al. 2011; Kengne et al. 2014).
The main limitation of the HealthImpact model is its reliance on claims data, such that to be subject to in silico screening, individuals must either be insured or have had some contact with the health care system to generate enrollment and billing information. This is not unique to models using administrative data, and current laboratory‐based methods require even greater access to and utilization of health care resources.
We could not directly compare the HealthImpact model with laboratory‐based prediabetes diagnostic criteria, as only a subset of our population had laboratory studies performed and available during the study period. People with laboratory data were different from the general population, as evidenced by their higher age, greater inclusion of minorities, and higher level of comorbidity. These characteristics also suggest greater risk for diabetes and morbidity than the general population, reinforcing the fact that laboratory studies are not equally likely to be obtained by all people at risk, and relying on them for screening purposes may miss a substantial number of people. In contrast, the HealthImpact model can simultaneously and without extra resource utilization screen large populations to identify those at risk.
Claims‐based screening strategies can be efficiently integrated into clinical practice. Different score thresholds can be used depending on the clinical context and goal of HealthImpact use. Specifically, higher score thresholds (>90) can be used to identify patients at very high risk who may not be currently in contact with the health care system, and proactively reach out to them to schedule a clinical encounter and diagnostic testing. A high PPV is necessary for cost‐effective outreach efforts and HealthImpact score >90 has PPV of 22 percent. Intermediate thresholds (>75) can identify patients who are already receiving ongoing but unrelated clinical care, but would also benefit from diabetes screening. In this case, a lower PPV of 17 percent may be cost‐effective. Finally, low thresholds (>50) could be used to flag patients as being at risk for diabetes and in whom the diagnosis of diabetes should be entertained if presenting with a constellation of clinically related symptoms or conditions. These patients may not be sought out proactively, justifying an even lower PPV of 10 percent. Health care providers, public health systems, and payers may have different objectives and available resources for identifying high‐risk patients (Linnan et al. 2008; Kottke et al. 2009; Duru et al. 2013; CDC 2015), and each could use a different HealthImpact score threshold that is specific to the population and task at hand. Moreover, personalized risk information can be directly transmitted to the patient and the health care team, facilitating not only identification and contact but also informed and shared decision making.
HealthImpact can be adapted to all health plans and systems that bill for or codify their services. While many health systems may initially lack the technological capacity or commitment to run the HealthImpact model, private/public health policy can be informed by monitoring deidentified patient populations for those at risk for diabetes. And with additional efforts, similar HealthImpact models could be developed for other conditions or constellations of conditions, including cardiovascular disease, hypertension, hyperlipidemia, and obesity.
Supporting information
Appendix SA1: Author Matrix.
Box S1. Objective of the Study and HealthImpact Development.
Box S2. Justification and Limitations of Nested Case–Control Study Design.
Box S3. Index Date for Controls in the Development and Internal Validation Datasets.
Box S4. Independent Variables.
Box S5. Model Fitting.
Box S6. Clinical Vignettes of HealthImpact Implementation.
Box S7. Timeliness of Diabetes Risk Predictions in External Validation Dataset.
Box S8. C‐Statistic in Testing Data.
Box S9. HealthImpact versus Laboratory‐Based Risk Stratification (Sensitivity Analysis).
Figure S1. Sparsity of Subjects in the Training Dataset by Percentiles of Total Number of Available Nondemographic Variables.
Figure S2. Receiver Operating Curve (ROC) for Training and Internal Validation Data.
Figure S3. Time to Diabetes Diagnosis in the External Validation Dataset Stratified by Baseline HealthImpact Score.
Table S1. Baseline Characteristics of People in the Incident Diabetes and Control Cohorts Included in the HealthImpact Training and Internal Validation Populations.
Table S2. HealthImpact Accuracy in the Training Population as a Function of Baseline Score.
Table S3. Kaplan–Meier Estimates of Type 2 Diabetes Incidence at One‐, Two‐, and Three‐Year Intervals.
Table S4. Baseline Characteristics of People Included in the Overall HealthImpact External Validation Cohort and in the Subset Used in Sensitivity Analysis Comparing HealthImpact to Laboratory‐Based Definitions of Prediabetes.
Table S5. Comparison of HealthImpact and Laboratory Methods for Predicting Risk of Incident Diabetes at One, Two, and Three Years in a Subset of the External Validation Dataset Where Laboratory Data Were Available.
Acknowledgments
Joint Acknowledgment/Disclosure Statement: V. Nori and C. Hane received salary support for developing this paper from Optum Labs. Funding for this study came from Optum Labs. Optum Labs had no role in the design and conduct of the study; collection, management, analysis, and interpretation of data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. All conclusions are those of the authors alone, and none of the authors declare any potential conflicts of interest.
Disclosures: None.
Disclaimers: None.
References
- Abbasi, A. , Peelen L. M., Corpeleijn E., van der Schouw Y. T., Stolk R. P., Spijkerman A. M. W., van der A D. L., Moons K. G. M., Navis G., Bakker S. J. L., and Beulens J. W. J.. 2012. “Prediction Models for Risk of Developing Type 2 Diabetes: Systematic Literature Search and Independent External Validation Study.” BMJ 345: e5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann, R. T. , Cheng Y. J., Williamson D. F., and Gregg E. W.. 2011. “Identifying Adults at High Risk for Diabetes and Cardiovascular Disease Using Hemoglobin A1c: National Health and Nutrition Examination Survey 2005–2006.” American Journal of Preventive Medicine 40 (1): 11–7. [DOI] [PubMed] [Google Scholar]
- ADA . 2015. “American Diabetes Association. Standards of Medical Care in Diabetes—2015.” Diabetes Care 38 (Suppl. 1): S8–S16. [Google Scholar]
- Balkau, B. , Lange C., Fezeu L., Tichet J., de Lauzon‐Guillain B., Czernichow S., F. Fumeron , Froguel P., Vaxillaire M., Cauchi S., Ducimetiere P., and E. Eschwege . 2008. “Predicting Diabetes: Clinical, Biological, and Genetic Approaches: Data from the Epidemiological Study on the Insulin Resistance Syndrome (DESIR).” Diabetes Care 31 (10): 2056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2011. Centers for Disease Control and Prevention. National Diabetes Fact Sheet: National Estimates and General Information on Diabetes and Prediabetes in the United States. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. [Google Scholar]
- CDC . 2012. “Annual Number (in Thousands) of New Cases of Diagnosed Diabetes Among Adults Aged 18–79 Years, United States, 1980–2011” [accessed on October 3, 2015]. Available at http://www.cdc.gov/diabetes/statistics/incidence/fig1.htm
- CDC . 2013. “Awareness of Prediabetes–United States, 2005‐2010.” MMWR Morbidity and Mortality Weekly Report 62 (11): 209–12. [PMC free article] [PubMed] [Google Scholar]
- CDC . 2015. “National Diabetes Prevention Program” [accessed on October 3, 2015]. Available at http://www.cdc.gov/diabetes/prevention/
- Chen, L. , Magliano D. J., Balkau B., Colagiuri S., Zimmet P. Z., Tonkin A. M., Mitchell P., Phillips P. J., and Shaw J. E.. 2010. “AUSDRISK: An Australian Type 2 Diabetes Risk Assessment Tool Based on Demographic, Lifestyle and Simple Anthropometric Measures.” Medical Journal of Australia 192 (4): 197–202. [DOI] [PubMed] [Google Scholar]
- Chiasson, J. L. , Josse R. G., Gomis R., Hanefeld M., Karasik A., and Laakso M.. 2002. “Acarbose for Prevention of Type 2 Diabetes Mellitus: The STOP‐NIDDM Randomised Trial.” Lancet 359 (9323): 2072–7. [DOI] [PubMed] [Google Scholar]
- Collins, S. R. , Robertson R., Garber T., and Doty M. M.. 2013. Insuring the Future: Current Trends in Health Coverage and the Effects of Implementing the Affordable Care Act. Findings from the Commonwealth Fund Biennial Health Insurance Survey, 2012. Commonwealth Fund pub. no. 1681. The Commonwealth Fund. [Google Scholar]
- Duru, O. K. , Mangione C. M., Chan C., Keckhafer A., Kimbro L., Kirvan K. A., Turk N., Luchs R., Li J., and Ettner S.. 2013. “Evaluation of the Diabetes Health Plan to Improve Diabetes Care and Prevention.” Preventing Chronic Disease 10: E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelgau, M. M. , Narayan K. M., and Herman W. H.. 2000. “Screening for Type 2 Diabetes.” Diabetes Care 23 (10): 1563–80. [DOI] [PubMed] [Google Scholar]
- Friedman, J. , Hastie T., and Tibshirani R.. 2010. “Regularization Paths for Generalized Linear Models via Coordinate Descent.” Journal of Statistical Software 33 (1): 1–22. [PMC free article] [PubMed] [Google Scholar]
- Garber, A. J. , Abrahamson M. J., Barzilay J. I., Blonde L., Bloomgarden Z. T., Bush M. A., Dagogo‐Jack S., Davidson M., Einhorn D., Garber J. R., Garvey W. T., Grunberger G., Handelsman Y., Hirsch I. B., Jellinger P. S., McGill J. B., Mechanick J. I., Rosenblit P. D., Umpierrez G., and Davidson M. H.. 2015. “AACE Comprehensive Diabetes Management Algorithm 2015.” Endocrine Practice 21(4): 438–47. [DOI] [PubMed] [Google Scholar]
- Harrell, F. J. 2001. Regression Modeling Strategies. New York: Springer. [Google Scholar]
- Harrell, F. E. 2014. “rms: Regression Modeling Strategies. R Package Version 4.2‐0” [accessed on October 3, 2015]. Available at http://CRAN.R-project.org/package=rms
- Herman, W. H. , Edelstein S. L., Ratner R., Montez M. G., Ackermann R. T., Orchard T., Foulkes M. A., Zhang P., Saudek C., and Brown M. B.. 2012. “The 10‐Year Cost‐Effectiveness of Lifestyle Intervention or Metformin for Diabetes Prevention: An Intent‐to‐Treat Analysis of the DPP/DPPOS.” Diabetes Care 35 (4): 723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, A. K. , Murty M. N., and Flynn P. J.. 1999. “Data Clustering: A Review.” ACM Computing Surveys 31 (3): 264–323. [Google Scholar]
- Kengne, A. P. , Beulens J. W. J., Peelen L. M., Moons K. G. M., van der Schouw Y. T., Schulze M. B., Spijkerman A. M. W., Griffin S. J., Grobbee D. E., Palla L., Tormo M.‐J., Arriola L., Barengo N. C., Barricarte A., Boeing H., Bonet C., Clavel‐Chapelon F., Dartois L., Fagherazzi G., Franks P. W., Huerta J. M., Kaaks R., Key T. J., Khaw K. T., Li K., Mühlenbruch K., Nilsson P. M., Overvad K., Overvad T. F., Palli D., Panico S., Quirós J. R., Rolandsson O., Roswall N., Sacerdote C., Sánchez M.‐J., Slimani N., Tagliabue G., Tjønneland A., Tumino R., van der A D. L., Forouhi N. G., Sharp S. J., Langenberg C., Riboli E., and Wareham N. J.. 2014. “Non‐Invasive Risk Scores for Prediction of Type 2 Diabetes (EPIC‐InterAct): A Validation of Existing Models.” Lancet Diabetes & Endocrinology 2 (1): 19–29. [DOI] [PubMed] [Google Scholar]
- King, G. , and Zeng L.. 2001. “Logistic Regression in Rare Events Data.” Political Analysis 9 (2): 137–63. [Google Scholar]
- Knowler, W. C. , Barrett‐Connor E., Fowler S. E., Hamman R. F., Lachin J. M., Walker E. A., and Nathan D. M.. 2002. “Reduction in the Incidence of Type 2 Diabetes with Lifestyle Intervention or Metformin.” New England Journal of Medicine 346 (6): 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler, W. C. , Fowler S. E., Hamman R. F., Christophi C. A., Hoffman H. J., Brenneman A. T., Brown‐Friday J. O., Goldberg R., Venditti E., and Nathan D. M.. 2009. “10‐Year Follow‐Up of Diabetes Incidence and Weight Loss in the Diabetes Prevention Program Outcomes Study.” Lancet 374 (9702): 1677–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottke, T. E. , Jordan C. O., O'Connor P. J., Pronk N. P., and Carreon R.. 2009. “Readiness of US Health Plans to Manage Cardiometabolic Risk.” Preventing Chronic Disease 6 (3): A86. [PMC free article] [PubMed] [Google Scholar]
- Krieger, N. , Chen J. T., Waterman P. D., Rehkopf D. H., and Subramanian S. V.. 2003. “Race/Ethnicity, Gender, and Monitoring Socioeconomic Gradients in Health: A Comparison of Area‐Based Socioeconomic Measures–The Public Health Disparities Geocoding Project.” American Journal of Public Health 93 (10): 1655–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström, J. , and Tuomilehto J.. 2003. “The Diabetes Risk Score: A Practical Tool to Predict Type 2 Diabetes Risk.” Diabetes Care 26 (3): 725–31. [DOI] [PubMed] [Google Scholar]
- Linnan, L. , Bowling M., Childress J., Lindsay G., Blakey C., Pronk S., Wieker S., and Royall P.. 2008. “Results of the 2004 National Worksite Health Promotion Survey.” American Journal of Public Health 98 (8): 1503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCQA . 2009. “HEDIS 2009 Volume 2 Technical Update.” National Committee for Quality Assurance (NCQA) [accessed on November 11, 2014]. Available at http://www.ncqa.org/portals/0/PolicyUpdates/HEDIS%20Technical%20Updates/09_CDC_Spec.pdf [Google Scholar]
- Nichols, G. A. , and Brown J. B.. 2005. “Higher Medical Care Costs Accompany Impaired Fasting Glucose.” Diabetes Care 28 (9): 2223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, D. , Mathur R., Dent T., Meads C., and Greenhalgh T.. 2011. “Risk Models and Scores for Type 2 Diabetes: Systematic Review.” BMJ 343: d7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Office of the National Coordinator for Health Information Technology (ONC) within the Office of the Secretary for the U.S. Department of Health and Human Services (HHS) . 2013a. “Compatibility & Information Exchange. Health Information Exchange” [accessed on October 3, 2015]. Available at http://www.healthit.gov/providers-professionals/health-information-exchange
- Optum . 2014. “Optum Labs Data Warehouse Technical Specifications” [accessed on August 18, 2015]. Available at https://www.optum.com/content/dam/optum/resources/productSheets/5302_Data_Assets_Chart_Sheet_ISPOR.pdf
- Pan, X. R. , Li G. W., Hu Y. H., Wang J. X., Yang W. Y., An Z. X., Hu Z. X., Lin J., Xiao J. Z., Cao H. B., Liu P. A., Jiang X. G., Jiang Y. Y., Wang J. P., Zheng H., Zhang H., Bennett P. H., and Howard B. V.. 1997. “Effects of Diet and Exercise in Preventing NIDDM in People with Impaired Glucose Tolerance: The Da Qing IGT and Diabetes Study.” Diabetes Care 20 (4): 537–44. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . 2013b. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; [accessed on October 3, 2015]. Available at http://www.R-project.org/ [Google Scholar]
- Sacks, D. B. 2011. “A1C versus Glucose Testing: A Comparison.” Diabetes Care 34 (2): 518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, M. B. , Hoffmann K., Boeing H., Linseisen J., Rohrmann S., Möhlig M., Pfeiffer A. F. H., Spranger J., Thamer C., Häring H.‐U., Fritsche A., and Joost H.‐G.. 2007. “An Accurate Risk Score Based on Anthropometric, Dietary, and Lifestyle Factors to Predict the Development of Type 2 Diabetes.” Diabetes Care 30 (3): 510–5. [DOI] [PubMed] [Google Scholar]
- Steyerberg, E. 2009. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. New York: Springer. [Google Scholar]
- Tabaei, B. P. , Burke R., Constance A., Hare J., May‐Aldrich G., Parker S. A., Scott A., Stys A., Chickering J., and Herman W. H.. 2003. “Community‐Based Screening for Diabetes in Michigan.” Diabetes Care 26 (3): 668–70. [DOI] [PubMed] [Google Scholar]
- Tuomilehto, J. , Lindstrom J., Eriksson J. G., Valle T. T., Hamalainen H., Ilanne‐Parikka P., Keinanen‐Kiukaanniemi S., Laakso M., Louheranta A., Rastas M., Salminen V., and Uusitupa M.. 2001. “Prevention of Type 2 Diabetes Mellitus by Changes in Lifestyle among Subjects with Impaired Glucose Tolerance.” New England Journal of Medicine 344 (18): 1343–50. [DOI] [PubMed] [Google Scholar]
- Wallace, P. J. , Shah N. D., Dennen T., Bleicher P. A., and Crown W. H.. 2014. “Optum Labs: Building a Novel Node in the Learning Health Care System.” Health Affairs 33 (7): 1187–94. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Gregg E. W., Williamson D. F., Barker L. E., Thomas W., Bullard K. M., G. Imperatore , Williams D. E., and Albright A. L.. 2010. “A1C Level and Future Risk of Diabetes: A Systematic Review.” Diabetes Care 33 (7): 1665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix SA1: Author Matrix.
Box S1. Objective of the Study and HealthImpact Development.
Box S2. Justification and Limitations of Nested Case–Control Study Design.
Box S3. Index Date for Controls in the Development and Internal Validation Datasets.
Box S4. Independent Variables.
Box S5. Model Fitting.
Box S6. Clinical Vignettes of HealthImpact Implementation.
Box S7. Timeliness of Diabetes Risk Predictions in External Validation Dataset.
Box S8. C‐Statistic in Testing Data.
Box S9. HealthImpact versus Laboratory‐Based Risk Stratification (Sensitivity Analysis).
Figure S1. Sparsity of Subjects in the Training Dataset by Percentiles of Total Number of Available Nondemographic Variables.
Figure S2. Receiver Operating Curve (ROC) for Training and Internal Validation Data.
Figure S3. Time to Diabetes Diagnosis in the External Validation Dataset Stratified by Baseline HealthImpact Score.
Table S1. Baseline Characteristics of People in the Incident Diabetes and Control Cohorts Included in the HealthImpact Training and Internal Validation Populations.
Table S2. HealthImpact Accuracy in the Training Population as a Function of Baseline Score.
Table S3. Kaplan–Meier Estimates of Type 2 Diabetes Incidence at One‐, Two‐, and Three‐Year Intervals.
Table S4. Baseline Characteristics of People Included in the Overall HealthImpact External Validation Cohort and in the Subset Used in Sensitivity Analysis Comparing HealthImpact to Laboratory‐Based Definitions of Prediabetes.
Table S5. Comparison of HealthImpact and Laboratory Methods for Predicting Risk of Incident Diabetes at One, Two, and Three Years in a Subset of the External Validation Dataset Where Laboratory Data Were Available.


