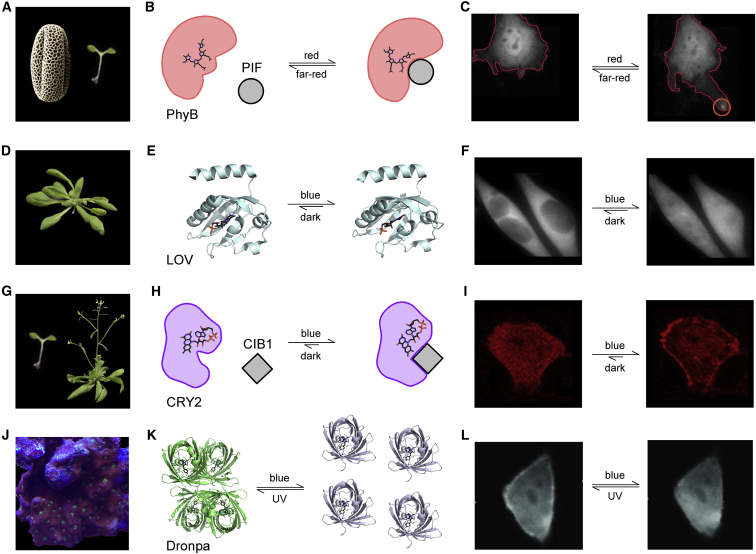
Figure 1.
Organism of origin and relevant lifecycle is shown for various optogenetic actuators. (A) PhyB is known to be involved in germination and flowering of Arabidopsis thaliana (84). (B) Cartoon depiction of PhyB/PIF photoswitching mechanism with chromophore structure (85). (de-etioliation C) PhyB/PIF-induced lamellipodia formation (red circle) (86). (D) LOV domain of phototropin is a key component of phototropism and stomatal opening in Arabidopsis (84). (E) Atomic structures show two light-dependent configurations of LOV domain isolated from Avena sativa (left, Protein Data Bank (PDB): 2V0U; right, PDB: 2V0W) without the Jα helix (87). An alternate mechanism is LOV photodimerization with an EL222 mutation and VVD. (F) Lov2-Jα mediated nuclear localization (88). (G) CRY2 is an important player in flowering and deetiolation of Arabidopsis (84). (H) Cartoon rendition of CRY2/CIB1 dimerization with blue light (89). Light-dependent oligomerization of CRY2 is another powerful optogenetic mode of operation. (I) Membrane localization of CRY2-Akt in response to blue light (44). (J) Dronpa is a homolog of GFP found in coral Pectiniidae. (K) Crystal structures show reverse dimerization of Dronpa (left, tetramer PDB: 2POX (32); right, monomers PDB: 2IOV (90)). (Green) Fluorescent state; (gray) dark state of Dronpa. (L) Functional exemplar of light-regulated photoswitching of Dronpa (30). For visualization of relocalization, the fluorescence of mNeptune tagged to Dronpa is monitored. Images in (A), (D), and (G) are reproduced from Krämer (91). To see this figure in color, go online.