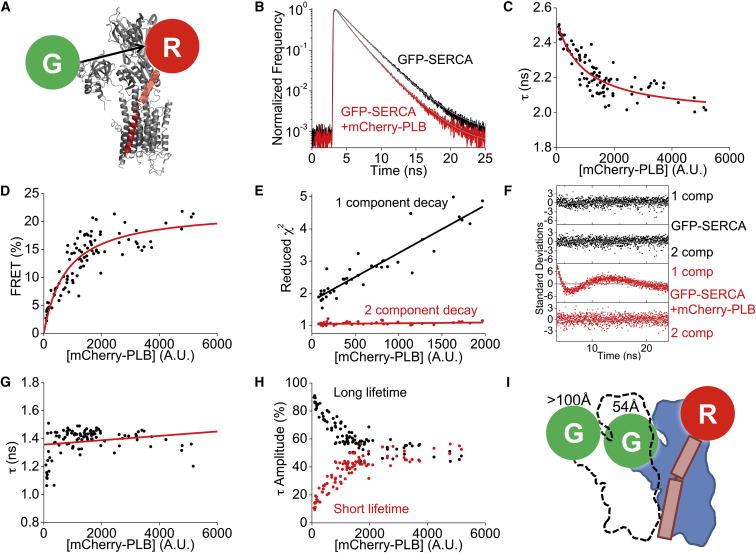
Figure 3.
Fluorescence-lifetime FRET analysis of the SERCA-PLB regulatory complex. (A) Labeling strategy for FRET from GFP-SERCA (gray) to mCherry-PLB (red). (B) Representative fluorescence decay of GFP-SERCA shortened by coexpression of mCherry-PLB, indicating FRET. Exponential tail fitting is shown in gray. (C) A single-component exponential fit of the fluorescence lifetime shows a decrease in τ with increasing protein expression. The hyperbolic model is in red. (D) FRET (calculated from the data in (C)) increased with protein expression. The hyperbolic model is in red. (E) Reduced-χ2 values for one-component fits worsened as protein expression increased. The reduced χ2 value was improved by a two-component fit. Linear fits appear as lines. (F) Residual plots of (B) from one- or two-component exponential decay fitting of the GFP-SERCA alone (black) or in the presence of mCherry-PLB (red). (G) The short-lifetime values from two-component exponential decays were consistent across a large range of protein expression levels. The linear fit is shown in red. (H) The relative contributions (amplitude) of the short and long lifetimes. (I) A model of the SERCA-SERCA-PLB regulatory complex in which one of the donor-labeled SERCA protomers (dotted outline) is too distant to participate in FRET with PLB. To see this figure in color, go online.