Fig. 2.
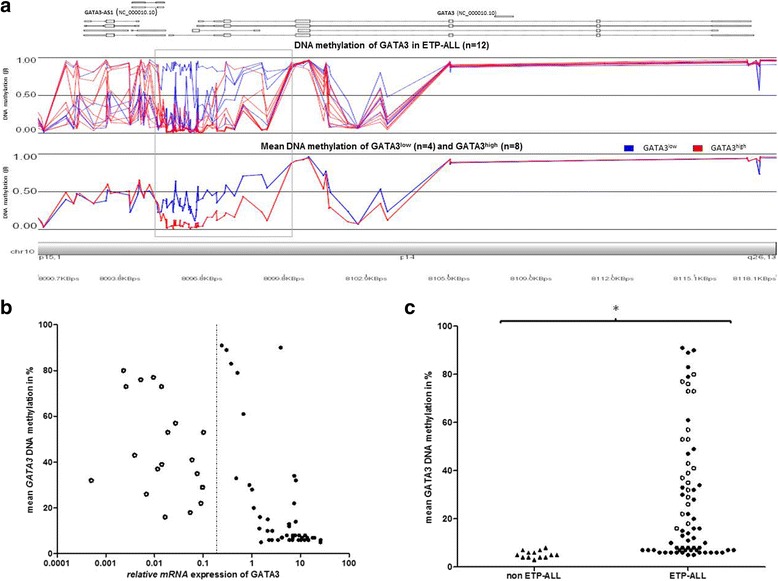
GATA3 silencing is regulated by DNA methylation in ETP-ALL. a 10p15 chromosome plot depicting DNA methylation of 12 ETP-ALL patient samples as assessed by Illumina Infinium® HumanMethylation450 BeadChip. β values representing DNA methylation for each patient (upper panel) and mean DNA methylation (lower panel) of GATA3low ETP-ALL (n = 4, blue) and GATA3high ETP-ALL (n = 8, red). Comparing GATA3low ETP-ALL (n = 4, blue) with GATA3high ETP-ALL (n = 8, red), 35 differentially methylated sites were located within a 6-kb segment of GATA3 (indicated by the gray box), including the CpGs that were analyzed by pyrosequencing in a larger cohort of patients. b GATA3 DNA methylation as assessed by pyrosequencing (of CpGs within the gray box in a) was negatively correlated to GATA3 mRNA expression in ETP-ALL (n = 64, r = −0.73, p < 0.0001). The dotted line indicates the cutoff to distinguish GATA3low (empty dots) and GATA3high (solid dots) samples. c Pyrosequencing revealed higher GATA3 DNA methylation in ETP-ALL (n = 69) than in non-ETP-ALL (n = 13) (28 vs. 5 %, p < 0.0001 indicated by asterisk). Empty and solid dots indicate GATA3low and GATA3high ETP-ALL, while triangles indicate non-ETP-ALL
