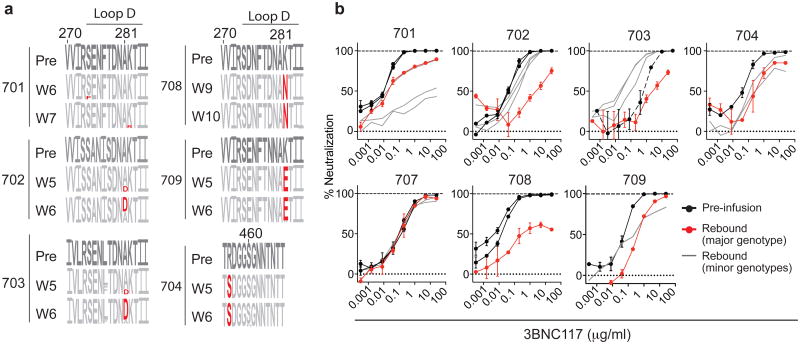
Figure 4. 3BNC117 resistance in rebound viruses.
a, Logogram shows env gp120 regions (amino acid positions; 270–285 and 455–467, according to HXBc2 numbering) indicating sequence changes from pre-infusion culture(s) (first row) to rebound sequences derived from plasma SGS at the indicated time points. The frequency of each amino acid is indicated by its height. Red residues represent mutations predicted to affect neutralization30. b, 3BNC117 neutralization sensitivity of pseudoviruses derived from pre-infusion or rebound SGS. Black lines represent pre-infusion virus envs; red lines represent the major env at rebound for each participant (Extended Data Figs 7 and 8, Supplementary Tables 8 and 9 and Methods); grey lines represent minor rebound envs in participants with multiple rebound viruses or variants that evolved after rebound (Extended Data Figs 7 and 8, Supplementary Table 9). Symbols reflect the means of two technical replicates; error bars denote standard deviation.