Abstract
The catalytic core of the self-splicing group I intron RNAs is composed of six paired regions together with their connecting sequences; these are thought to form two elongated domains, with paired regions P5, P4, and P6 aligned along one axis and P8, P3, and P7 along the other. Most of the very highly conserved residues of the group I introns lie in or near P7, but two occur in L4, the internal loop connecting P4 and P5. It is generally believed that such bases are conserved because they are essential for splicing. Mutants were created in a member of each of the two major subclasses of group I introns, in which P5, L4, and the distal portion of P4 were deleted. Splicing activity was still detected in these mutants, albeit substantially weakened; splicing was accurate and occurred by the normal group I mechanism, with addition of a guanosine molecule to the intron. Thus the deleted region, containing two universally conserved bases, is not essential but facilitates splicing. Another reaction characteristic of group I introns, hydrolysis of the 3' splice site, was less severely affected by the deletions. The results are discussed in terms of the prevailing three-dimensional model for the core structure of the group I introns.
Full text
PDF
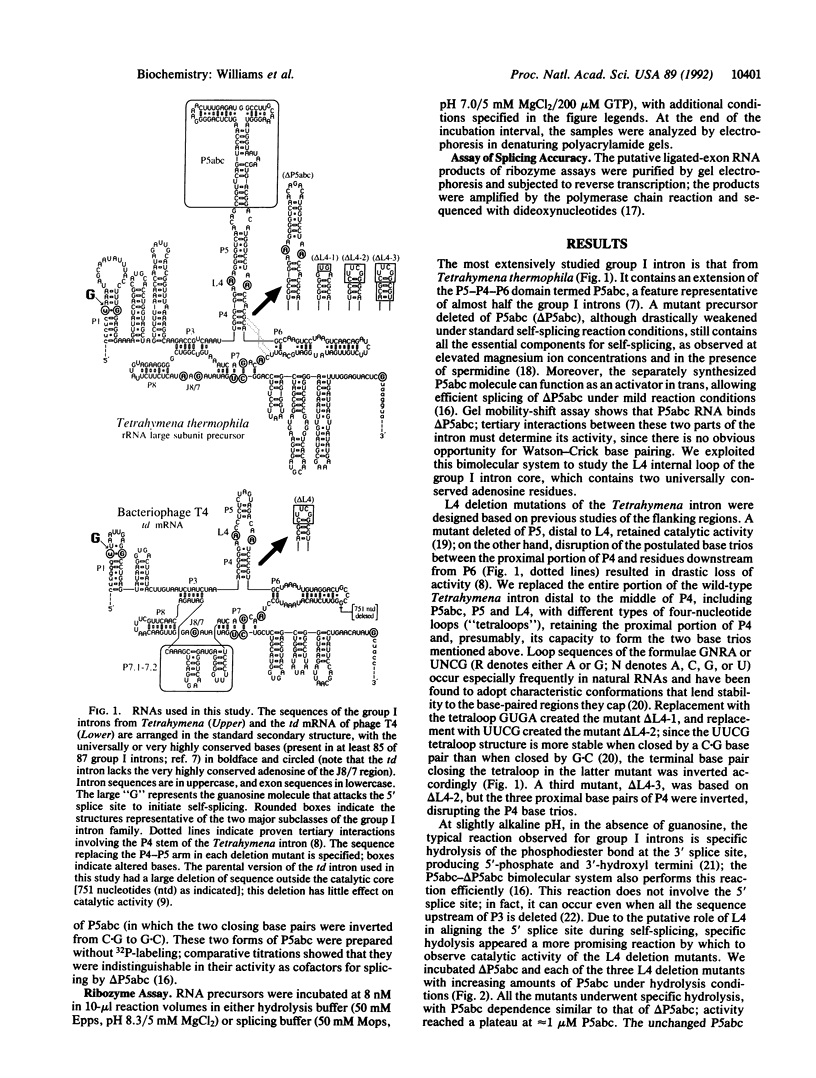
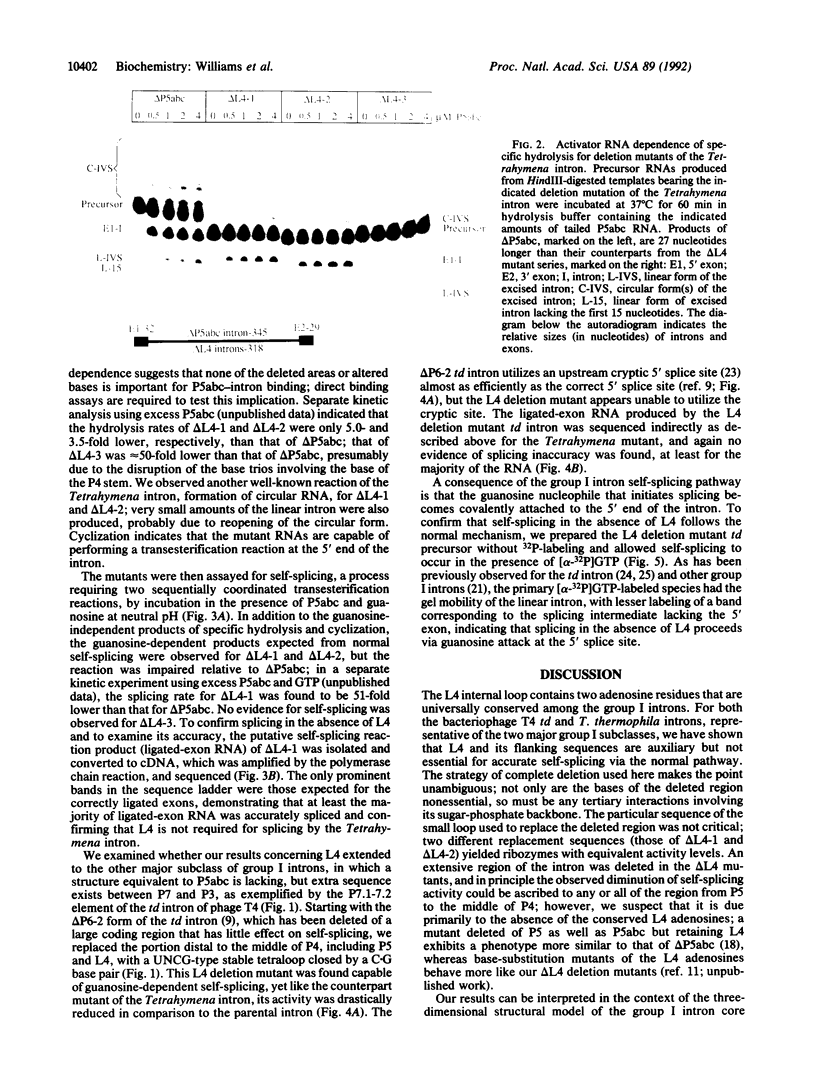
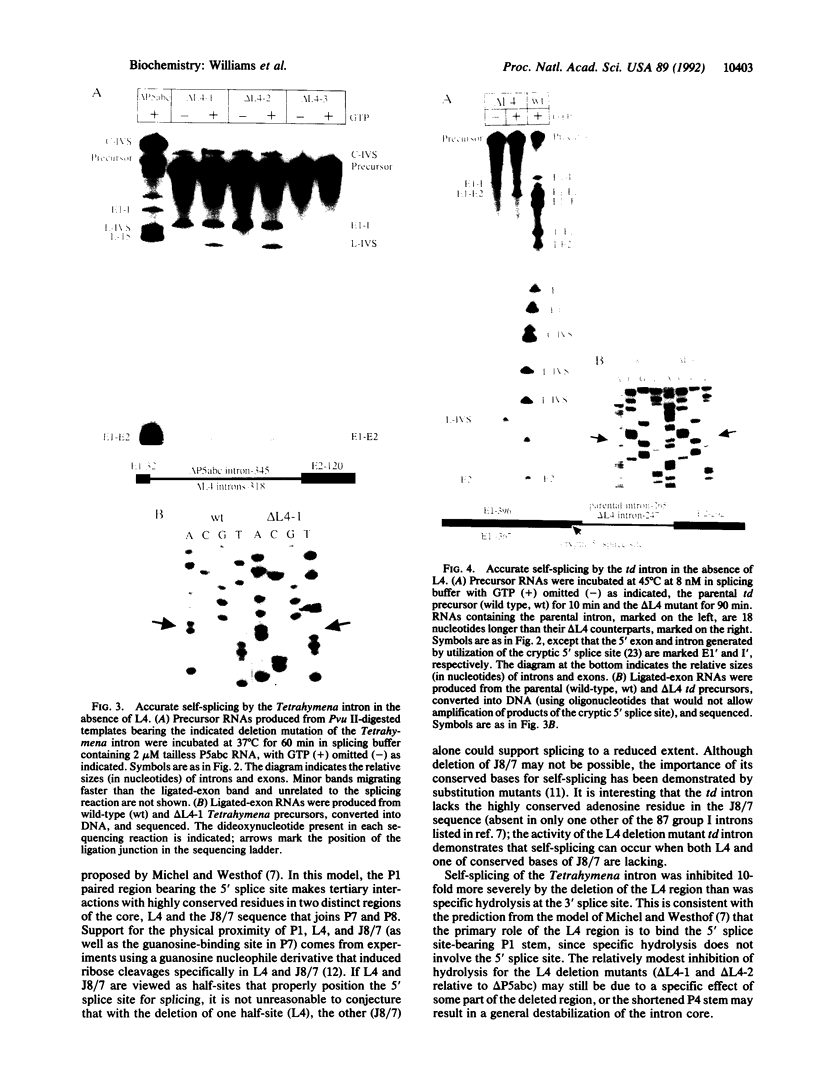

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antao V. P., Lai S. Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991 Nov 11;19(21):5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudry A. A., Joyce G. F. Minimum secondary structure requirements for catalytic activity of a self-splicing group I intron. Biochemistry. 1990 Jul 10;29(27):6534–6539. doi: 10.1021/bi00479a027. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986 Jan 31;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Chandry P. S., Belfort M. Activation of a cryptic 5' splice site in the upstream exon of the phage T4 td transcript: exon context, missplicing, and mRNA deletion in a fidelity mutant. Genes Dev. 1987 Nov;1(9):1028–1037. doi: 10.1101/gad.1.9.1028. [DOI] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., Maley F. Mechanism and requirements of in vitro RNA splicing of the primary transcript from the T4 bacteriophage thymidylate synthase gene. Biochemistry. 1987 Jun 2;26(11):3050–3057. doi: 10.1021/bi00385a016. [DOI] [PubMed] [Google Scholar]
- Couture S., Ellington A. D., Gerber A. S., Cherry J. M., Doudna J. A., Green R., Hanna M., Pace U., Rajagopal J., Szostak J. W. Mutational analysis of conserved nucleotides in a self-splicing group I intron. J Mol Biol. 1990 Oct 5;215(3):345–358. doi: 10.1016/s0022-2836(05)80356-0. [DOI] [PubMed] [Google Scholar]
- Doudna J. A., Szostak J. W. RNA-catalysed synthesis of complementary-strand RNA. Nature. 1989 Jun 15;339(6225):519–522. doi: 10.1038/339519a0. [DOI] [PubMed] [Google Scholar]
- Galloway Salvo J. L., Coetzee T., Belfort M. Deletion-tolerance and trans-splicing of the bacteriophage T4 td intron. Analysis of the P6-L6a region. J Mol Biol. 1990 Feb 5;211(3):537–549. doi: 10.1016/0022-2836(90)90264-m. [DOI] [PubMed] [Google Scholar]
- Gott J. M., Shub D. A., Belfort M. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell. 1986 Oct 10;47(1):81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Hemsley A., Arnheim N., Toney M. D., Cortopassi G., Galas D. J. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989 Aug 25;17(16):6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Sullivan F. X., Cech T. R. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J Mol Biol. 1986 May 5;189(1):143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- Joyce G. F., van der Horst G., Inoue T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989 Oct 11;17(19):7879–7889. doi: 10.1093/nar/17.19.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Cech T. R. Three-dimensional model of the active site of the self-splicing rRNA precursor of Tetrahymena. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8788–8792. doi: 10.1073/pnas.84.24.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Ellington A. D., Couture S., Szostak J. W. Phylogenetic and genetic evidence for base-triples in the catalytic domain of group I introns. Nature. 1990 Oct 11;347(6293):578–580. doi: 10.1038/347578a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns--a review. Gene. 1989 Oct 15;82(1):5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. Splicing of messenger RNA precursors. Science. 1987 Feb 13;235(4790):766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- Wang J. F., Cech T. R. Tertiary structure around the guanosine-binding site of the Tetrahymena ribozyme. Science. 1992 Apr 24;256(5056):526–529. doi: 10.1126/science.1315076. [DOI] [PubMed] [Google Scholar]
- Williamson C. L., Desai N. M., Burke J. M. Compensatory mutations demonstrate that P8 and P6 are RNA secondary structure elements important for processing of a group I intron. Nucleic Acids Res. 1989 Jan 25;17(2):675–689. doi: 10.1093/nar/17.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A. J., Groot Koerkamp M. J., Tabak H. F. The mechanism of group I self-splicing: an internal guide sequence can be provided in trans. EMBO J. 1990 Jun;9(6):1923–1928. doi: 10.1002/j.1460-2075.1990.tb08319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst G., Christian A., Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]






