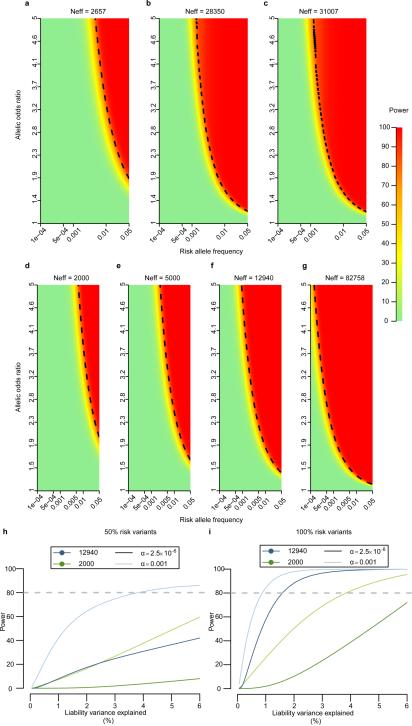
Extended Data Figure 4. Power for single and aggregate variant association.
a-g. Power to detect single-variant association (α=5×10−8) at varying minor allele frequency (x-axis) and allelic odds-ratio (y-axis) for seven effective sample size (Neff) scenarios relevant to the genomes (a-c) and exomes (dg) component of this project. a. variant observed in 2,657 samples (the effective size of the GoT2D integrated panel); b. variant observed in 28,350 samples (the effective size of the imputed data set); c. variant observed in the GoT2D integrated panel and the imputed data set (effective sample size 31,007); d. ancestry-specific variant in 2,000 samples (the size of each of the non-European exome sequence data sets); e. European specific variant in 5,000 samples (the combined size of the European exome sequence data sets); f. variant observed with shared frequency across all ancestry groups in 12,940 samples (the size of the combined exome sequence data set); and g. variant observed in the combined exome array and sequencing data set (effective sample size 82,758). h-i. Power for gene based test of association (SKAT-O) according to liability variance explained. In h, 50% of the variants contribute to disease risk while the remaining 50% have no effect on disease risk; in i., 100% of the variants contribute to disease risk. For each, sample sizes considered are 2,000 (ancestry-specific effects; green) and 12,940 (ancestry-shared effects; blue). Power is shown for two levels of significance (α=2.5×10−6 and α=0.001). From these simulation studies, it is clear that under the optimistic model, where effects are shared across all ethnicities (blue line) and all variants contribute, power is >60% for 1% variance explained and α=2.5×10−6. However, power declines rapidly if either criterion is relaxed.