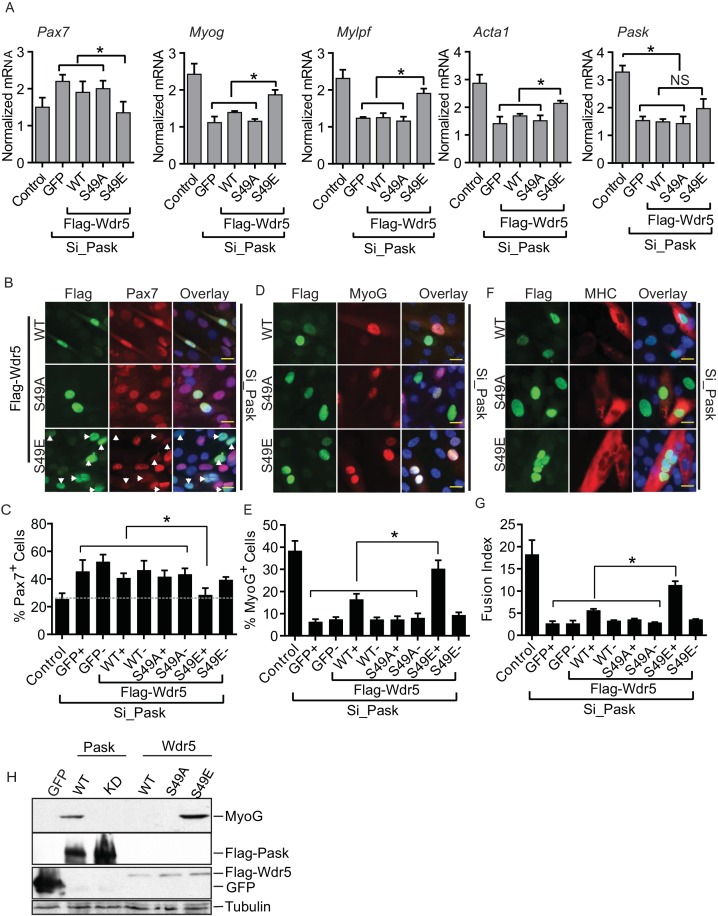
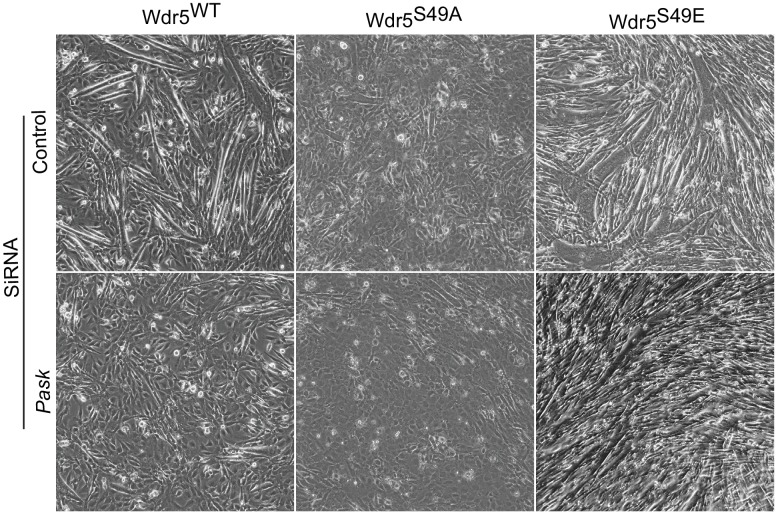
Figure 5. The phospho-mimetic S49E Wdr5 mutant rescues myogenesis in Pask-silenced cells.
(A) GFP or WT, S49A or S49E Wdr5 were retrovirally expressed in Pask-siRNA C2C12 cells. qRT-PCR analysis was performed for the indicated mRNA on day 3 of differentiation. 18S rRNA was used as normalizer. n = 3. Error bars ± S.D *p<0.05. (B) Flag-tagged WT, S49A or S49E Wdr5 was expressed in Pask-siRNA C2C12 cells. After 24 hr, cells were stained for Pax7 at Day 0 of differentiation. Arrows show Wdr5S49E-expressing cells and the corresponding cell autonomous decrease in Pax7 expression. Scale bar = 20 µM. (C) Quantification of percent Pax7+ cells from (B) as a function of the presence (+) or absence (−) of GFP or Wdr5. n = 3 independent experiments each with 100 cells counted. Error bars ± S.D. *p<0.05. (D) As in (B), except cells were stained for MyoG on Day 1 of differentiation. (E) Quantification of percent MyoG+ cells from (D) as a function of the presence (+) or absence (−) of GFP or Wdr5. n = 3 independent experiments each with 100 cells counted. Error bars ± S.D. *p<0.05. (F) As in (B), except cells were stained for MHC on Day 3 of differentiation. (G) Quantification of fusion index from (F) as a function of the presence (+) or absence (−) of GFP or Wdr5. n = 3 independent experiments each with 100 cells counted. Error bars ± S.D. *p<0.05. (H) C2C12 myoblasts were infected with retrovirus expressing GFP, Flag tagged WT or KD Pask or WT, S49A or S49E Wdr5 and infected cells were selected with puromycin in growth media for 48 hr. Cells were lysed after selection and abundance of the indicated proteins was determined by Western blotting.
DOI: http://dx.doi.org/10.7554/eLife.17985.018