Abstract
Microbacterium oleivorans is a predominant member of hydrocarbon-contaminated environments. We here report on the genomic analysis of M. oleivorans strain Wellendorf that was isolated from an indoor door handle. The partial genome of M. oleivorans strain Wellendorf consists of 2,916,870 bp of DNA with 2831 protein-coding genes and 49 RNA genes. The organism appears to be a versatile mesophilic heterotroph potentially capable of hydrolysis a suite of carbohydrates and amino acids. Genomic analysis revealed metabolic versatility with genes involved in the metabolism and transport of glucose, fructose, rhamnose, galactose, xylose, arabinose, alanine, aspartate, asparagine, glutamate, serine, glycine, threonine and cysteine. This is the first detailed analysis of a Microbacterium oleivorans genome.
Keywords: Microbacterium oleivorans, Draft genome, Detailed annotation, Student Initiated Microbial Discovery (SIMD) project, Bioremediation potential, Metabolic versatility
1. Introduction
The strain Wellendorf was isolated from a door handle surface with frequent human use in Stillwater, OK as part of the Student Initiated Microbial Discovery (SIMD) project (introduced in [1]). The Microbacterium genus is a phylogenetically and physiologically diverse genus with members ubiquitously found in polycyclic aromatic hydrocarbon (PAH)-contaminated [2], [3], as well as heavy metal-contaminated [4], [5] soils. PAHs and heavy metals are persistent environmental contaminants with both environmental and human health concerns [6], [7], [8]. Genomic analysis of strains belonging to the genus Microbacterium can contribute to our understanding of the molecular mechanisms of PAHs degradation and heavy metal mobilization and could potentially contribute to natural-attenuation-based, and engineered bioremediation schemes in multiple environments [9], [10]. Here we present the draft genomic sequence, and first detailed genomic annotation and analysis of a Microbacterium oleivorans strain.
2. Materials and methods
2.1. Genome project history
The draft assembly and annotation were completed in 2015–2016. Table 1 shows the genome project information.
Table 1.
Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Draft |
| MIGS-28 | Libraries used | 2 × 300 paired end chemistry |
| MIGS 29 | Sequencing platforms | Illumina Miseq |
| MIGS 31.2 | Fold coverage | 300 × |
| MIGS 30 | Assemblers | Velvet 2.0 |
| MIGS 32 | Gene calling method | Prodigal |
| GenBank ID | MAYO00000000 | |
| GenBank date of release | July 2016 | |
| GOLD ID | Gp0126761 | |
| BIOPROJECT | PRJNA327390 | |
| MIGS 13 | Project relevance | Environmental |
2.2. Growth conditions and genomic DNA preparation
M. oleivorans strain Wellendorf was grown overnight at 30 °C on tryptic soy agar plates. Genomic DNA of high sequencing quality was isolated using the MPBio PowerSoil® DNA extraction kit according to manufacturer's instructions. Negative stain TEM micrographs were obtained using the services of the Oklahoma State University Microscopy Lab. Briefly, the sample was placed on a carbon film TEM grid and allowed to incubate for 2 min, after which the excess liquid was wicked off. Phosphotungestic acid (PTA; 2% w/v) was then added to the grid followed by a 45-s incubation. Excess PTA was blotted off and the grid was allowed to dry before it was visualized using JOEL JEM-2100 transmission electron microscope.
2.3. Genome sequencing and assembly
The genome of M. oleivorans strain Wellendorf was sequenced using the Illumina MiSeq platform at the University of Georgia Genomics Facility using 2 × 300 paired end chemistry and an average library insert size of 700 bp. Quality filtered sequence data were assembled with the short read de Bruijn graph assembly program Velvet [11] using a kmer value of 101 bp and a minimum contig coverage value of 7 ×. The genome project is deposited in GOLD (Genomes On-Line Database) and this Whole Genome Shotgun (WGS) project has been deposited in GenBank under the accession MAYO00000000. The version described in this paper is version MAYO01000000.
2.4. Genome annotation
Gene models were created using the prokaryotic gene calling software package Prodigal [12]. A total of 2885 gene models were predicted. The average gene size was 961 bp. Translated protein sequences were functionally annotated using a combination of NCBI Blast C ++ homology search and HMMER 3.0 [13] hmmscan against the PFAM 26.0 database [14]. Additional gene analysis and functional annotation were carried out through the Integrated Microbial Genomes Expert Review (IMG-ER) platform.
2.5. Phylogenetic analysis
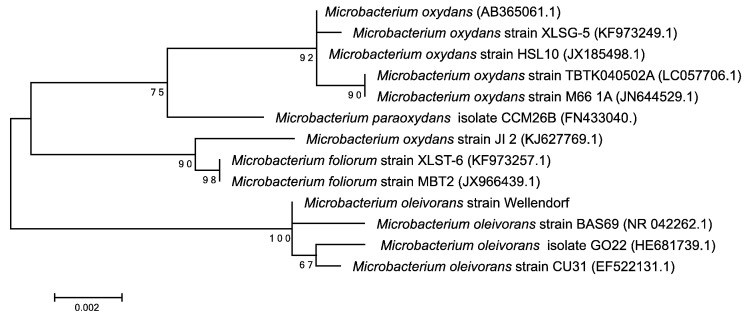
A maximum likelihood phylogenetic tree was constructed using multiple sequence alignments of 16S rRNA genes sequences. Multiple sequence alignment was conducted in Mega, as were the selection of the best substitution model, and the maximum likelihood analysis [15]. The tree was obtained under “TN93 + G + I” model with, a proportion of invariable sites of 0.25, and a variable site γ shape parameter of 0.51. Escherichia coli partial 16S rRNA gene isolate ECSD9 was used as the outgroup. Bootstrap values, in percent, were based on 200 replicates.
2.6. Comparative genomics
We sought to compare the genome of Microbacterium oleivorans strain Wellendorf to 17 closely related genomes (IMG genome IDs: 2576861779, 2519899511, 2639762631, 2627854169, 2619619265, 2609459760, 2576861795, 2639762630, 2636415545, 2645728100, 2540341240, 2643221903, 2627854213, 2541047020, 2608642165, 2522572100, and 2526164566) using the “Genome clustering” function on the IMG-ER analysis platform based on the COG profile. We also used principal component analysis to compare the genomes based on several genomic features including the genome size, the number of genes, the number of transporters identified, the GC content, the number of non-coding bases, the number of genes belonging to COG categories, as well as the number of genes belonging to each COG category [16], [17]. The PCA analysis was conducted using the “princomp” function in the labdsv library of R [18]. The results were visualized using a biplot, where genomes were represented by stars and genomic features or COG categories used for comparison were represented by arrows, where the arrow directions follow the maximal abundance, and their lengths are proportional to the maximal rate of change between samples.
3. Results and discussion
3.1. Classification and features
Cells of M. oleivorans strain Wellendorf are Gram positive, non-motile, aerobic irregular rods that were arranged in pairs (Fig. 1). Colonies on TSA agar were orange-red.
Fig. 1.
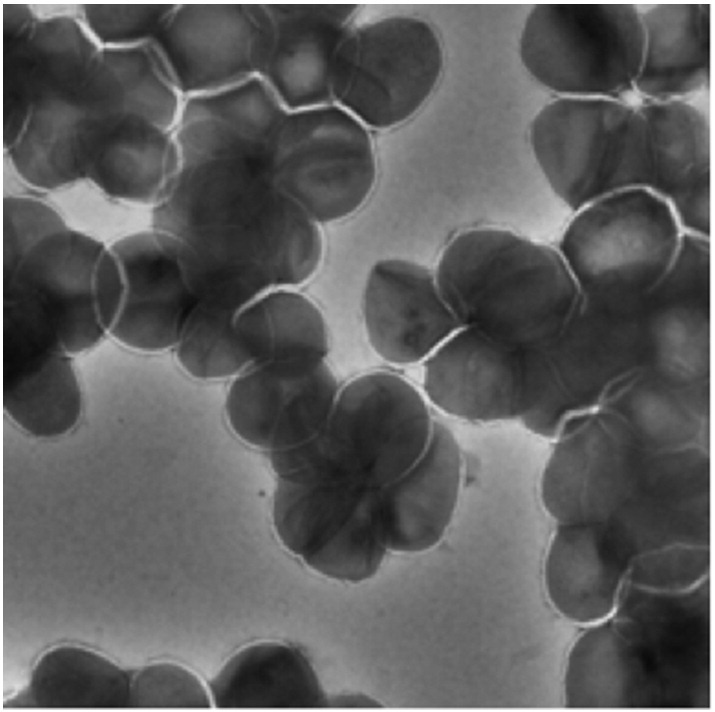
Negative stain TEM micrograph of Microbacterium oleivorans strain Wellendorf.
Within the genus Microbacterium, 94 species are described with validly published names. Strain Wellendorf shares 93.23–100% 16S rRNA gene identity with other species in the Microbacterium genus (Table 2). Compared to other Microbacterium oleivorans strains with sequenced genomes, Strain Wellendorf shares 99% 16S rRNA gene similarity with Microbacterium oleivorans strains CD11_3 (GenBank accession number LSTV00000000) and NBRC 103075 (GenBank accession number BCRG01000000), and 100% similarity to strain RIT293 [19].
Table 2.
M. oleivorans strain Wellendorf 16S rRNA gene percentage similarity to other Microbacterium species.
| Microbacterium species | Type strain | Wellendorf strain % similarity |
|---|---|---|
| M. aerolatum | V-73 | 98.27% |
| M. agarici | CC-SBCK-209 | 94.05% |
| M. amylolyticum | N5 | 93.23% |
| M. aoyamense | KV-492 | 97.82% |
| M. aquimaris | JS54-2 | 97.97% |
| M. arabinogalactanolyticum | ATCC 51926 | 97.44% |
| M. arborescens | ATCC 4358 | 97.20% |
| M. arthrosphaerae | CCM 7681 | 97.48% |
| M. aurantiacum | ATCC 49090 | 97.89% |
| M. aurum | ATCC 51345 | 97.51% |
| M. awajiense | YM13-414 | 97.66% |
| M. azadirachtae | AI-S262 | 97.95% |
| M. binotii | CIP 101303 | 97.42% |
| M. chocolatum | BUCSAV 207 | 97.72% |
| M. deminutum | KV-483 | 97.66% |
| M. dextranolyticum | M-73 | 97.89% |
| M. enclense | NIO-1002 | 97.81% |
| M. endophyticum | PA15 | 97.35% |
| M. esteraromaticum | ATCC 8091 | 97.51% |
| M. flavescens | ATCC 13348 | 98.12% |
| M. flavum | YM18-098 | 98.58% |
| M. fluvii | YSL3-15 | 97.89% |
| M. foliorum | P 333/02 | 98.43% |
| M. ginsengisoli | Gsoil 259 | 96.35% |
| M. ginsengiterrae | DCY37 | 98.65% |
| M. gubbeenense | DPC 5286 | 93.25% |
| M. halimionae | PA36 | 97.58% |
| M. halophilum | N° 76 | 96.02% |
| M. halotolerans | YIM 70130 | 95.63% |
| M. hatanonis | FCC-01 | 97.51% |
| M. hominis | CIP 105731 | 98.27% |
| M. humi | CC-12309 | 94.12% |
| M. hydrocarbonoxydans | BNP48 | 98.35% |
| M. hydrothermale | 0704C9-2 | 97.58% |
| M. immunditiarum | SK 18 | 96.50% |
| M. imperial | ATCC 8365 | 97.28% |
| M. indicum | BBH6 | 94.53% |
| M. insulae | DS-66 | 98.12% |
| M. invictum | DSM 19600 | 97.51% |
| M. jejuense | THG-C31 | 97.20% |
| M. keratanolyticum | ATCC 35057 | 98.42% |
| M. ketosireducens | CIP 105732 | 97.66% |
| M. kitamiense | C2 | 97.81% |
| M. koreense | JS53-2 | 97.74% |
| M. kribbense | MSL-04 | 95.88% |
| M. kyungheense | THG-C26 | 97.82% |
| M. lacticum | ATCC 8180 | 98.12% |
| M. lacus | A5E-52 | 97.67% |
| M. laevaniformans | ATCC 15953 | 97.88% |
| M. lemovicicum | ViU22 | 97.74% |
| M. lindanitolerans | MNA2 | 93.77% |
| M. luteolum | ATCC 51474 | 98.34% |
| M. luticocti | SC-087B | 94.99% |
| M. mangrove | MUSC 115 | 96.82% |
| M. marinilacus | YM11-607 | 95.80% |
| M. marinum | H101 | 98.04% |
| M. maritypicum | ATCC 19260 | 98.42% |
| M. mitrae | M4-8 | 97.04% |
| M. murale | 01-Gi-001 | 97.88% |
| M. nanhaiense | OAct400 | 93.98% |
| M. natoriense | TNJL143-2 | 98.80% |
| M. neimengense | 7087 | 97.20% |
| M. oleivorans | BAS69 | 100% |
| M. oryzae | MB10 | 95.64% |
| M. oxydans | DSM 20578 | 98.42% |
| M. paludicola | US15 | 95.57% |
| M. panaciterrae | DCY56 | 97.67% |
| M. paraoxydans | CF36 | 98.73% |
| M. petrolearium | LAM0410 | 96.81% |
| M. phyllosphaerae | P 369/06 | 98.65% |
| M. populi | 10-107-8 | 94.67% |
| M. profundi | Shh49 | 98.12% |
| M. proteolyticum | RZ36 | 97.97% |
| M. pseudoresistens | CC-5209 | 96.66% |
| M. pumilum | KV-488 | 97.74% |
| M. pygmaeum | KV-490 | 97.67% |
| M. radiodurans | GIMN 1.002 | 97.43% |
| M. rhizomatis | DCY100 | 95.17% |
| M. saccharophilum | K-1 | 98.04% |
| M. saperdae | ATCC 19272 | 98.27% |
| M. schleiferi | ATCC 51473 | 98.42% |
| M. sediminicola | YM10-847 | 96.81% |
| M. sediminis | YLB-01 | 96.27% |
| M. shaanxiense | CCNWSP60 | 97.90% |
| M. soli | DCY 17 | 95.25% |
| M. suwonense | M1T8B9 | 96.27% |
| M. terrae | ATCC 51476 | 97.65% |
| M. terregens | ATCC 13345 | 97.74% |
| M. terricola | KV-448 | 97.74% |
| M. thalassium | CIP 105728 | 98.12% |
| M. trichothecenolyticum | ATCC 51475 | 97.82% |
| M. ulmi | XIL02 | 96.66% |
| M. xylanilyticum | S3-E | 97.05% |
| M. yannicii | G72 | 97.89% |
Phylogenetic analysis based on the 16S rRNA gene placed strain M. oleivorans BAS69 as the closest taxonomic relative of M. oleivorans strain Wellendorf (Table 3 and Fig. 2).
Table 3.
Classification and general features of M. oleivorans strain Wellendorf [30].
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [22] | |
| Phylum Actinobacteria | TAS [22] | ||
| Class Actinobacteria | TAS [22] | ||
| Order Micrococcales | TAS [22] | ||
| Family Microbacteriaceae | TAS [22] | ||
| Genus Microbacterium | TAS [22] | ||
| Species oleivorans | TAS [22] | ||
| (Type) strain: Wellendorf | |||
| Gram stain | Positive | TAS [22] | |
| Cell shape | Irregular rods | TAS [22] | |
| Motility | Non-motile | TAS [22] | |
| Sporulation | Non-spore forming | TAS [22] | |
| Temperature range | Mesophile | TAS [22] | |
| Optimum temperature | 30 °C | TAS [22] | |
| pH range; optimum | Unknown | ||
| Carbon source | l-arabinose, d-cellobiose, d-fructose, d-galactose, gluconate, d-glucose, d-maltose, d-mannose, α-d-melibiose, l-rhamnose, d-ribose, d-sucrose, salicin, d-trehalose, l-xylose, d-mannitol, sorbitol, fumarate, dl-lactate, l-malate, pyruvate, l-aspartate, l-histidine, putrescine and 4-hydroxybenzoate | TAS [22] | |
| MIGS-6 | Habitat | Indoor environment, door handle | TAS [22] |
| MIGS-6.3 | Salinity | 2–4% NaCl (w/v) | TAS [22] |
| MIGS-22 | Oxygen requirement | Obligate aerobe | TAS [22] |
| MIGS-15 | Biotic relationship | free-living | IDA |
| MIGS-14 | Pathogenicity | Unknown | |
| MIGS-4 | Geographic location | USA | IDA |
| MIGS-5 | Sample collection | March 2016 | IDA |
| MIGS-4.1 | Latitude | 36.1157 | IDA |
| MIGS-4.2 | Longitude | − 97.0586 | IDA |
| MIGS-4.4 | Altitude | 1 M | IDA |
Evidence codes - IDA: inferred from direct assay; TAS: traceable author statement (i.e., a direct report exists in the literature); NAS: non-traceable author statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [31].
Fig. 2.
A maximum likelihood phylogenetic tree constructed using multiple sequence alignments of 16S rRNA genes. “Microbacterium oleivorans strain Wellendorf” sequence is shown in bold. GenBank accession numbers are given in parentheses. The tree was obtained under “TN93 + G + I” model with, a proportion of invariable sites of 0.25, and a variable site γ shape parameter of 0.51. The tree was rooted using Escherichia coli partial 16S rRNA gene isolate ECSD9 (not shown). Bootstrap values, in percent, are based on 200 replicates and are shown for branches with > 50% bootstrap support. Multiple sequence alignment, model selection, and maximum likelihood analysis using MEGA [15].
3.2. Genome properties
The genome assembly produced a contig N50 of 2,860,671 bp with a total genome size of 2,916,870 bp. The GC content was 69.57%. Forty nine RNA genes were identified in the genome including 4 ribosomal RNA and 45 tRNA genes. The ribosomal RNA operon showed an atypical organization. Of the 2885 detected, 2831 were protein-coding, of which 76.26% had a function prediction, 65.34% represented a COG functional category, and 4.99% were predicted to have a signal peptide. Psort [20] classified proteins as 49.45% cytoplasmic, 0.85% extracellular, and 31.54% associated with the membrane. Based on the presence of 139 single copy genes [21], the genome is predicted to be 77.69% complete. Genome statistics are shown in Table 4. The distribution of genes into COG functional categories is shown in Table 5.
Table 4.
Genome statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 2,916,870 | 100% |
| DNA coding (bp) | 2,726,938 | 93.49% |
| DNA G + C (bp) | 2,029,207 | 69.57% |
| DNA scaffolds | 2 | 100% |
| Total genes | 2885 | 100% |
| Protein coding genes | 2831 | 98.13% |
| RNA genes | 54 | 1.87% |
| Pseudo genes | 0 | |
| Genes in internal clusters | 527 | 18.27% |
| Genes with function prediction | 2159 | 74.84% |
| Genes assigned to COGs | 1889 | 65.48% |
| Genes with Pfam domains | 2271 | 78.72% |
| Genes with signal peptides | 144 | 4.99% |
| Genes with transmembrane helices | 807 | 27.97% |
| CRISPR repeats | 0 |
Table 5.
Number of genes associated with general COG functional categories.
| Code | Value | % age | Description |
|---|---|---|---|
| J | 163 | 7.66% | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.05% | RNA processing and modification |
| K | 191 | 8.98% | Transcription |
| L | 96 | 4.51% | Replication, recombination and repair |
| B | 0 | 0% | Chromatin structure and dynamics |
| D | 22 | 1.03% | Cell cycle control, cell division, chromosome partitioning |
| V | 40 | 1.88% | Defense mechanisms |
| T | 88 | 4.14% | Signal transduction mechanisms |
| M | 98 | 4.61% | Cell wall/membrane biogenesis |
| N | 16 | 0.75% | Cell motility |
| U | 29 | 1.36% | Intracellular trafficking and secretion |
| O | 82 | 3.85% | Posttranslational modification, protein turnover, chaperones |
| C | 106 | 4.98% | Energy production and conversion |
| G | 230 | 10.81% | Carbohydrate transport and metabolism |
| E | 217 | 10.2% | Amino acid transport and metabolism |
| F | 76 | 3.75% | Nucleotide transport and metabolism |
| H | 123 | 5.78% | Coenzyme transport and metabolism |
| I | 93 | 4.37% | Lipid transport and metabolism |
| P | 108 | 5.08% | Inorganic ion transport and metabolism |
| Q | 38 | 1.79% | Secondary metabolites biosynthesis, transport and catabolism |
| R | 203 | 9.54% | General function prediction only |
| S | 95 | 4.46% | Function unknown |
| – | 1000 | 34.66% | Not in COGs |
The total is based on the total number of protein coding genes in the genome.
3.3. Insights from the genome sequence
Genome analysis of M. oleivorans strain Wellendorf identified a Gram positive microorganism with an atypical cell wall structure, with genomic evidences of a peptidoglycan layer lacking pentaglycine bridges and with meso-diaminopimelic acid (meso-DAP) as the second amino acid in the peptide linkage. This is different from Microbacterium oleivorans type strain whose cell wall was shown to be devoid of meso-DAP [22]. We identified genes encoding for the biosynthesis of the phosphoglycerolipid CDP-diacyl-glycerol in the genome. The analysis also revealed the absence of flagellar assembly genes and the presence of extracellular structures including Flp and Type IV pilus.
Further genomic analysis identified almost compete to complete catabolic KEGG pathways for each of the following carbon sources; glucose, fructose, rhamnose, galactose, xylose, arabinose, alanine, aspartate, asparagine, glutamate, serine, glycine, threonine and cysteine, and fatty acids as carbon and energy sources. The genome also encodes a complete TCA cycle and electron transport chain with P/V/-type ATPase subunits confirming the aerobic nature of the microorganism. While lactate and acetate fermentation capabilities were also identified in the genome, the facultative nature of this organism was not confirmed in the lab. Genomic analysis suggested auxotrophy for arginine, asparagine, thiamine, ubiquinone and biotin. In agreement with this observation, comparison of the protein-coding genes against the transporter database [23] identified several ABC and secondary transporters that could potentially import these elements.
When compared against the virulence factor database [24], the genome of M. oleivorans strain Wellendorf showed 668 virulence factor hits (19% of the protein-coding genes). These included secretion systems Type I and Type VII, among others.
The Wellendorf genome also encoded several proteins with bioremediation potential. These include enzymes for 4-hydroxyphenylacetate degradation via the meta-cleavage pathway, as well as for detoxification of nitronate [25], a known plant-secreted toxin [26], and of nitriloacetate [27], a chelating agent used in industry and frequently encountered in soil [28]. The genome also encodes for enzymes that can salvage S from organo-S-compounds (e.g. alkanesulfonates) in cases of limiting inorganic S [29].
3.4. Insights from comparative genomics
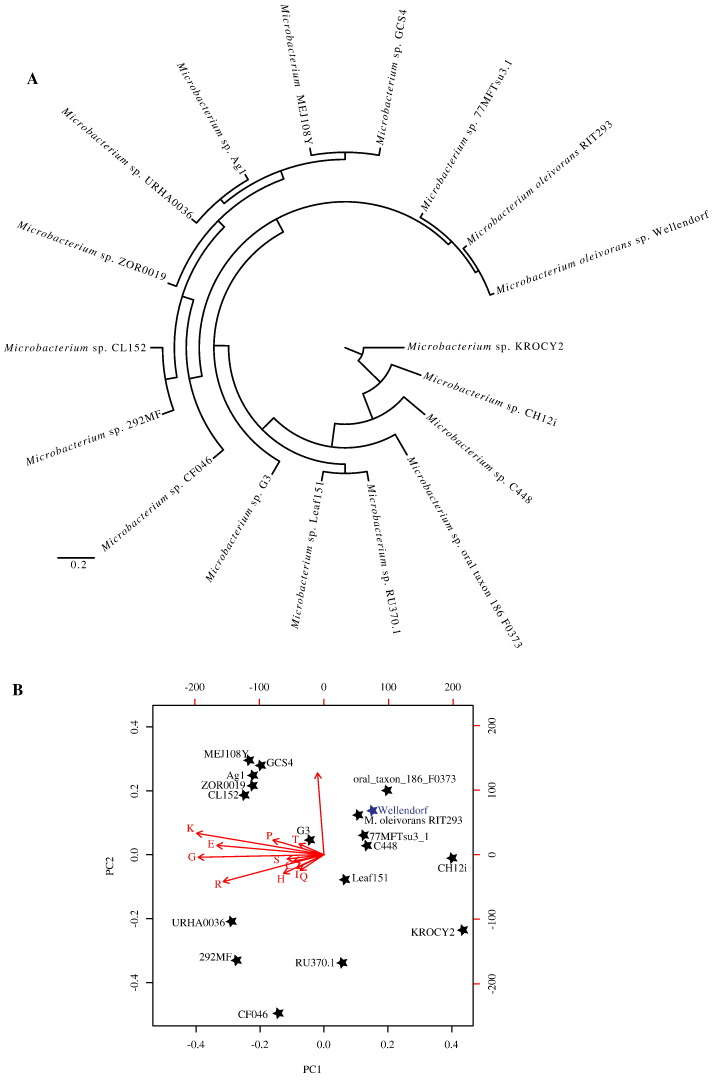
When the genome of M. oleivorans strain Wellendorf was compared to 17 closely related genomes based on their COG profile, the genome clustered with Microbacterium olievorans strain RIT293 (Fig. 3A). A closer look at the COG function profile of M. oleivorans strain Wellendorf in comparison to only Microbacterium oleivorans strains is shown in Table S1. Similarity to M. oleivorans strains at the functional level was in agreement with the phylogenetic position of the isolate as a member of the genus (Fig. 2). We used genomic features including the genome size, the number of genes, the number of transporters identified, the GC content, the number of non-coding bases, the number of genes belonging to COG categories, as well as the number of genes belonging to each COG category to cluster M. oleivorans strain Wellendorf genome in comparison to the 17 other closely related genomes. Results are shown in Fig. 3B. The genome of M. oleivorans strain Wellendorf clustered with the other M. oleivorans genome based on the enrichment in the number of transporters identified in the genomes.
Fig. 3.
(A) COG profile clustering of the genomes compared in this study. (B) Principal component analysis biplot based on the genomic features and COG category distribution in the genomes compared. Genomes are represented by stars (strain names are shown). Strain Wellendorf is shown in blue. Arrows represent genomic features or COG categories used for comparison. The arrow directions follow the maximal abundance, and their lengths are proportional to the maximal rate of change between genomes. The first two components explained 75% of variation.
4. Conclusions
This study presents the genome sequence and annotation of Microbacterium oleivorans strain Wellendorf. The genome revealed an extensive sugar and amino acid degradation machinery (for glucose, fructose, rhamnose, galactose, xylose, arabinose, alanine, aspartate, asparagine, glutamate, serine, glycine, threonine and cysteine). Comparison to the virulence factor database identified 668 genes in the genome with potential virulence-associated function including type Type I, and Type VII secretion systems. The genome also suggests the capability of degradation of fatty acid and the detoxification of several environmental contaminants including phenylacetate, nitronate, and nitriloacetate. Comparative genomics using general genomic features as well as the COG function profile coincided with the phylogenetic position predicted based on the 16S rRNA gene sequence and clustered the strain Wellendorf with another representative of the M. oleivorans species.
The following are the supplementary data related to this article.
Comparison of the COG function profile of strain Wellendorf and two other M. oleivorans strain. Only COG families with a representative in at least one of the three genomes are shown. Pearson correlations based on the abundances of the different COG families in the three genomes are shown below the table for all possible pairwise compaisons.
Transparency document
Transparency document.
Competing interests
All authors declare no competing interests.
Authors' contributions
APA, ELH, CL, MBC, and NY contributed to the analysis. APA, WDH, DPF, and NY wrote the manuscript. RW, CB, and RAH performed the lab experiments.
Acknowledgements and funding
Microbacterium oleivorans strain Wellendorf was selected for sequencing as part of a project at Oklahoma State University funded by the Howard Hughes Medical Institute aimed at improving student persistence through authentic, undergraduate research. The strain was isolated by an undergraduate student (RW) in an introductory microbiology course, modified to be the initial course in our microbial-discovery and genome-analysis two-semester course sequence. The genome was analyzed by a team of undergraduate (ALH and CL) and graduate (APA) students as part of an upper division microbial genomics class. This is Draft Genome #4 in the SIMD project supported in part by a grant from the Howard Hughes Medical Institute (grant number 1554854) through the Science Education Program. WDH acknowledges support by NSF grants MCB-1051590, MRI-1338097, and CHE-1412500.
Footnotes
The Transparency document associated with this article can be found, in the online version.
References
- 1.Couger M.B. Draft genome sequence of the environmental isolate Chryseobacterium sp. Hurlbut01. Genome Announc. 2015;3(5) doi: 10.1128/genomeA.01071-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaushik P. Arsenic hyper-tolerance in four Microbacterium species isolated from soil contaminated with textile effluent. Toxicol. Int. 2012;19(2):188–194. doi: 10.4103/0971-6580.97221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lal D. Microbacterium lindanitolerans sp. nov., isolated from hexachlorocyclohexane-contaminated soil. Int. J. Syst. Evol. Microbiol. 2010;60(Pt 11):2634–2638. doi: 10.1099/ijs.0.017699-0. [DOI] [PubMed] [Google Scholar]
- 4.Henson M.W. Metabolic and genomic analysis elucidates strain-level variation in Microbacterium spp. isolated from chromate contaminated sediment. PeerJ. 2015;3 doi: 10.7717/peerj.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corretto E. Draft genome sequences of 10 Microbacterium spp., with emphasis on heavy metal-contaminated environments. Genome Announc. 2015;3(3) doi: 10.1128/genomeA.00432-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chetwittayachan T., Shimazaki D., Yamamoto K. A comparison of temporal variation of particle-bound polycyclic aromatic hydrocarbons (pPAHs) concentration in different urban environments: Tokyo, Japan, and Bangkok, Thailand. Atmos. Environ. 2002;36(12):2027–2037. [Google Scholar]
- 7.Tchounwou P.B. Heavy metals toxicity and the environment. EXS. 2012;101:133–164. doi: 10.1007/978-3-7643-8340-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freije A.M. Heavy metal, trace element and petroleum hydrocarbon pollution in the Arabian Gulf: review. J. Assoc. Arab Univ. Basic Appl. Sci. 2015;17:90–100. [Google Scholar]
- 9.Abhilash P.C., Srivastava S., Singh N. Comparative bioremediation potential of four rhizospheric microbial species against lindane. Chemosphere. 2011;82(1):56–63. doi: 10.1016/j.chemosphere.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Pattanapipitpaisal P., Brown N.L., Macaskie L.E. Chromate reduction by Microbacterium liquefaciens immobilised in polyvinyl alcohol. Biotechnol. Lett. 2001;23(1):61–65. [Google Scholar]
- 11.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyatt D. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mistry J. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41(12) doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn R.D. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youssef N.H. Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum ‘Diapherotrites’. ISME J. 2015;9(2):447–460. doi: 10.1038/ismej.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youssef N.H., Ashlock-Savage K.N., Elshahed M.S. Phylogenetic diversities and community structure of members of the extremely halophilic Archaea (order Halobacteriales) in multiple saline sediment habitats. Appl. Environ. Microbiol. 2012;78(5):1332–1344. doi: 10.1128/AEM.07420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts D. Labdsv: ordination and multivariate analysis for ecology. 2007.
- 19.Gan H.Y. Whole-genome sequences of 13 endophytic bacteria isolated from shrub willow (Salix) grown in Geneva, New York. Genome Announc. 2014;2(3) doi: 10.1128/genomeA.00288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton P. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(Web Server issue):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinke C. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499(7459):431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 22.Schippers A. Microbacterium oleivorans sp. nov. and Microbacterium hydrocarbonoxydans sp. nov., novel crude-oil-degrading Gram-positive bacteria. Int. J. Syst. Evol. Microbiol. 2005;55(Pt 2):655–660. doi: 10.1099/ijs.0.63305-0. [DOI] [PubMed] [Google Scholar]
- 23.Saier M.H., Jr. The transporter classification database. Nucleic Acids Res. 2014;42(Database issue):D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33(Database issue):D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salvi F. The combined structural and kinetic characterization of a bacterial nitronate monooxygenase from Pseudomonas aeruginosa PAO1 establishes NMO class I and II. J. Biol. Chem. 2014;289(34):23764–23775. doi: 10.1074/jbc.M114.577791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis K. The biochemistry of the metabolic poison propionate 3-nitronate and its conjugate acid, 3-nitropropionate. IUBMB Life. 2013;65(9):759–768. doi: 10.1002/iub.1195. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y. Structure of nitrilotriacetate monooxygenase component B from Mycobacterium thermoresistibile. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67(Pt 9):1100–1105. doi: 10.1107/S1744309111012541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH: flavin mononucleotide oxidoreductase. J. Bacteriol. 1997;179(4):1112–1116. doi: 10.1128/jb.179.4.1112-1116.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis H.R. Mechanism for sulfur acquisition by the alkanesulfonate monooxygenase system. Bioorg. Chem. 2011;39(5-6):178–184. doi: 10.1016/j.bioorg.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Field D. The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the COG function profile of strain Wellendorf and two other M. oleivorans strain. Only COG families with a representative in at least one of the three genomes are shown. Pearson correlations based on the abundances of the different COG families in the three genomes are shown below the table for all possible pairwise compaisons.
Transparency document.