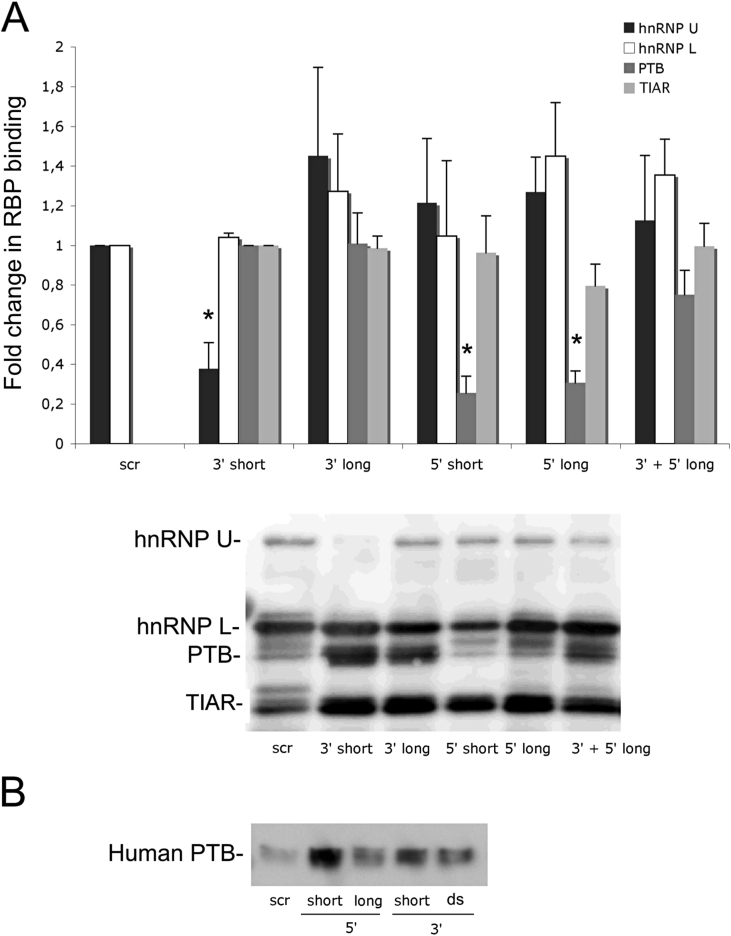
Fig. 3.
In vitro affinity of RNA binding proteins to oligonucleotides corresponding to specific 3′- and 5′-UTR sequences of rat (A) and human (B) insulin mRNA. RNA binding proteins from INS-1 cells were affinity purified using Dynabeads, separated by SDS-PAGE and analyzed by immunoblotting. The oligonucleotides used were those described in Fig. 2 and in one group (3′ + 5′ long) both the 3′ long and the 5′ long oligonucleotides were loaded on the beads so that complexes between the two oligonucleotides could be formed (Fig. 2B). In (A), protein bands for hnRNP U and hnRNP L were normalized against the signal from a scrambled control oligonucleotide (scr) while PTB and TIAR were normalized against the signal from the short oligonucleotide corresponding to the 3′-UTR of the rat insulin mRNA. The lower panel in A shows a representative blot with all four proteins. In (B), the PTB signal from a corresponding experiment using human islets is shown. Instead of a longer 3′-UTR oligonucleotide a non-biotinylated complementary oligonucleotide was used to mimic double stranded RNA. The blot is representative for two experiments. Results in A are means ± SEM for 4 independent experiments and * denotes p < 0.05 using Student’s paired t-test when comparing vs. corresponding control. Full images can be found in Supplemental Fig. 3.