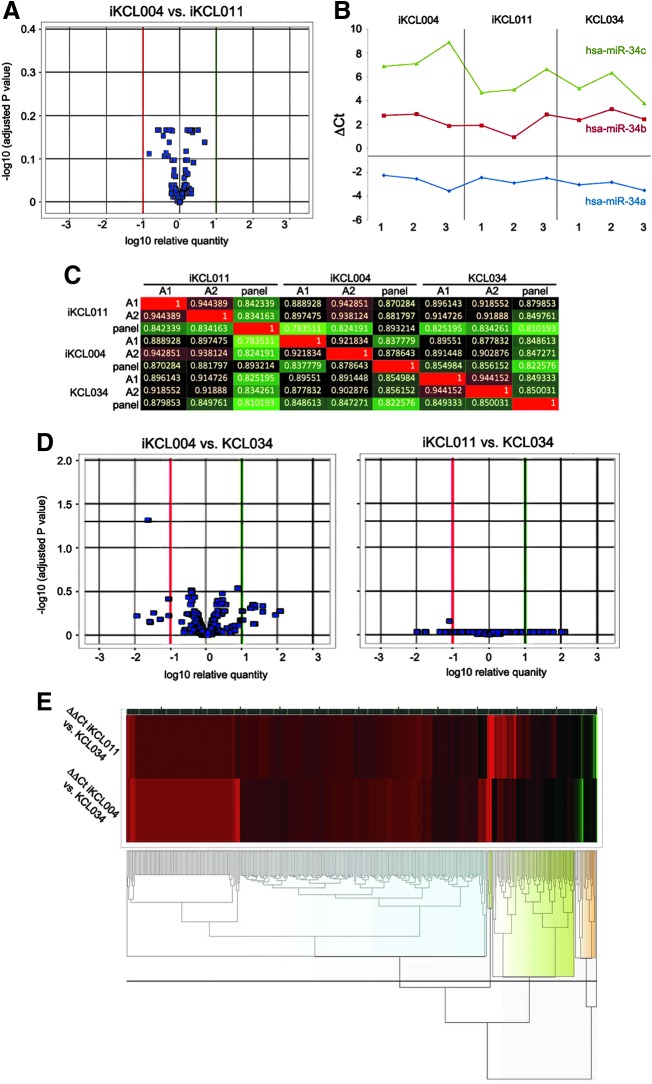
FIG. 2.
hESC/iPSC miRNA profiling. (A) Volcano plot showing no significant miRNA differentially expressed between two iPSC lines (n = 3 biological replicates per group). Significance versus fold change on the y- and x-axes, respectively, is plotted. Horizontal black lines represent the statistical significance threshold. Green and red lines represent the upper and lower fold changes thresholds, respectively. (B) Correlation heatmap among the profiles of miRNA expression of all the samples analyzed (three biological replicates per each of two iPSC and one hESC line). Superior to the heatmap and on the left of it three biological replicates and their relative grouping are reported. The color intensity varies in function of the Pearson correlation index ranging from green to red for a lower or a higher correlation, respectively. The proper numerical value of correlation between each couple of sample is reported within the squares at the intersections. (C) Volcano plot showing no significant miRNA differentially expressed between two iPSC lines, iKCL004 and iKCL011, and a control hESC line KCL034. (D) Hierarchical clustering of fold-change values obtained from the comparison of the two iPSC lines against hESC line used as a calibrator. The analysis was performed exploiting complete linkage, Euclidean distance, and z-score normalization, and the heatmaps, beside reporting four clusters of genes showing a similar behavior (highlighted by the four groups of dendograms with the same colored halo below the heatmaps), do not reveal significant differences among the samples analyzed. (E) Hierarchical clustering of fold-change values obtained from the comparison of the two iPSC lines against hESC line used as a calibrator. The analysis was performed exploiting complete linkage, Euclidean distance, and z-score normalization and the heatmaps, beside reporting four clusters of genes showing a similar behavior (highlighted by the four groups of dendograms with the same colored halo below the heatmaps), do not reveal significant differences among the samples analyzed. iPSC, induced pluripotent stem cell.