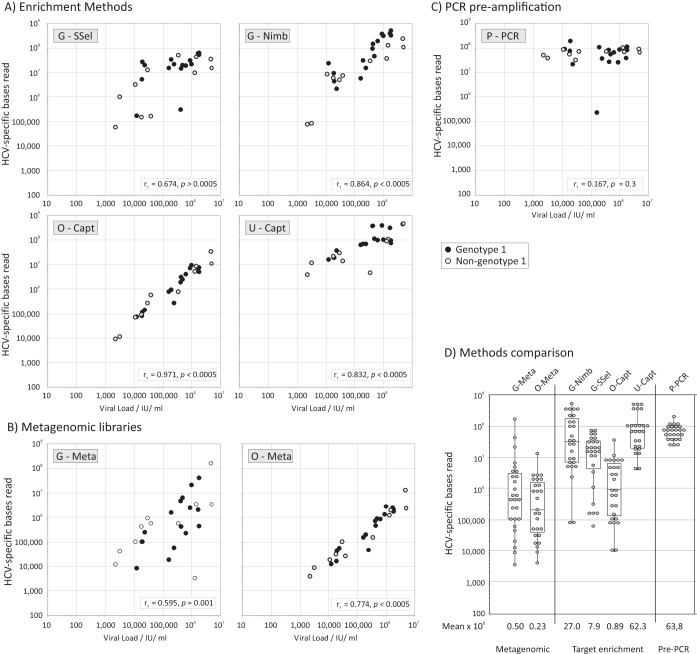
FIG 1.
Relationship between viral loads and read counts for each method. (A to C) Total HCV-specific bases read from each sample (y axis, log scale) was compared with viral loads separately for target enrichment (A), metagenomic library (B), and sequence preamplified by PCR (C), on a common x/y scale. Genotype 1 and non-genotype 1 samples are indicated according to the symbol key. The significance of the association between viral loads and read counts was calculated by Spearman's rank order correlation test; Spearman correlation coefficient (rs) values and P values are provided in inset boxes. (D) Distribution of viral loads by method with logarithmic mean values shown below the x axis. The box-and-whisker plots shows the median values and 67 and 95 percentiles.