Abstract
Infection is an important complication in patients with hematologic malignancies or solid tumors undergoing intensive cytotoxic chemotherapy. In only 20 to 30% of the febrile neutropenic episodes, an infectious agent is detected by conventional cultures. In this prospective study, the performance of broad-range PCR coupled with electrospray ionization time of flight mass spectrometry (PCR/ESI-MS) technology was compared to conventional blood cultures (BC) in a consecutive series of samples from high-risk hematology patients. In 74 patients, BC and a whole-blood sample for PCR/ESI-MS (Iridica BAC BSI; Abbott, Carlsbad, CA, USA) were collected at the start of each febrile neutropenic episode and, in case of persistent fever, also at day 5. During 100 different febrile episodes, 105 blood samples were collected and analyzed by PCR/ESI-MS. There was evidence of a bloodstream infection (BSI) in 36/105 cases (34%), based on 14 cases with both PCR/ESI-MS and BC positivity, 17 cases with BC positivity only, and 5 cases with PCR/ESI-MS positivity only. The sensitivity of PCR/ESI-MS was 45%, specificity was 93%, and the negative predictive value was 80% compared to blood culture. PCR/ESI-MS detected definite pathogens (Fusobacterium nucleatum and Streptococcus pneumoniae) missed by BC, whereas it missed both Gram-negative and Gram-positive organisms detected by BC. PCR/ESI-MS testing detected additional microorganisms but showed a low sensitivity (45%) compared to BC in neutropenic patients. Our results indicate a lower concordance between BC and PCR/ESI-MS in the neutropenic population than what has been previously reported in other patient groups with normal white blood cell distribution, and a lower sensitivity than other PCR-based methods.
INTRODUCTION
Infection is a leading cause of mortality in patients with hematologic malignancies or solid tumors, especially in those with prolonged and profound neutropenia (e.g., following remission induction or consolidation therapy for acute leukemia and high-risk myelodysplastic syndrome) and in recipients of myeloablative or reduced-intensity allogeneic stem cell transplantation (and, to a lesser extent, autologous stem cell transplant recipients) (1). Virtually all of these patients present with one or more episodes of fever during the neutropenic phase or during the posttransplant graft-versus-host disease (GvHD) period. Fever during these episodes is usually considered a sign of infection, triggering the immediate administration of broad-spectrum antimicrobial therapy. However, in only 20 to 30% of the febrile neutropenic episodes, an infectious agent is detected by the current gold standard for diagnosing bloodstream infections (BSI) (1, 2). This standard consists of continuous monitoring liquid blood culture (BC), followed by Gram stain, subculturing, identification, and antimicrobial susceptibility testing. However, the suboptimal sensitivity and long turnaround time of blood cultures (range, 2 to 5 days) are major shortcomings. In addition, fever is a highly nonspecific clinical sign. Hence, there is an urgent need for tools that can differentiate infectious from noninfectious fever and that can, in case of infectious fever, identify the underlying pathogen(s). Without the detection of an infectious agent, broad-spectrum antibiotics are often continued until recovery of neutrophils. A method with an excellent negative predictive value could potentially help reduce the inadequate use of antibiotics by ruling out BSI, while a high sensitivity might identify pathogens otherwise not detected by the blood culture systems.
During the last decade, several new technologies were developed and studied for diagnosis of BSI (3). These techniques focus on speeding up the identification of the microorganism(s) from blood cultures (e.g., use of matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS]) or focus on direct detection of the microorganism in blood samples. In 2008, an innovative technology based on a broad-range PCR coupled with electrospray ionization time of flight mass spectrometry (PCR/ESI-MS) technology was described for the detection and identification of microorganisms directly in clinical samples. The newest assays, including the BAC BSI assay (Ibis Biosciences, Abbott, Carlsbad, CA), were CE-IVD marked in 2015. Compared to the previous Ibis BSI assay, the Iridica BAC BSI has enhanced sensitivity, due to an increase in the blood volume tested, the optimization of PCR conditions and reagents to be tolerant of high loads of human DNA, and an improved downstream processing and analysis step (4). In two recently published prospective studies, PCR/ESI-MS provided rapid BSI pathogen detection and identification, with overall high sensitivity and negative predictive value (NPV) in intensive care unit (ICU) and emergency room (ER) patients (5, 6). In the ICU study by Vincent et al., PCR/ESI-MS was three times more likely to identify a pathogen than standard culture (5). In that regard, the early detection by PCR/ESI-MS of bacterial and fungal DNA in a blood sample, collected from a high-risk patient with fever, looks promising and might be able to overcome the current diagnostic shortcomings.
The aim of this study is to test the performance of the PCR/ESI-MS BAC BSI assay in a consecutive series of high-risk hematology patients and to compare its performance with that of conventional culture methods.
MATERIALS AND METHODS
Patient characteristics.
This prospective study was performed at the Acute Leukemia and Allogeneic Stem Cell Transplantation Unit of the University Hospitals Leuven (Leuven, Belgium) between September 2014 and April 2015. The study was approved by the local ethics committee. All patients provided written informed consent before any study-related procedure was performed. Patients undergoing remission induction or consolidation chemotherapy for acute leukemia (myeloid and lymphoid), allogeneic and autologous hematopoietic transplantation, or having prolonged neutropenia as a result of an underlying bone marrow disorder (e.g., aplastic anemia) were eligible as soon as they developed neutropenic fever. At the first episode of neutropenic fever, a single 10-ml EDTA sample for PCR/ESI-MS (BAC BSI/Iridica; Abbott, Carlsbad, CA, USA) was collected concomitantly with sets of blood cultures (from each lumen of the central indwelling line and a peripheral sample); additional samples (PCR and blood cultures) were collected at day 5 in patients with persistent neutropenic fever. Fever was defined as an oral temperature of ≥38.3°C in a single measurement. In all patients, antimicrobial prophylaxis consisted of fluconazole (400 mg daily) and levofloxacin (500 mg daily) given from the initiation of the chemotherapy or conditioning regimen until recovery of neutropenia or engraftment. In accordance with a diagnostic-driven approach for diagnosing invasive aspergillosis, serum galactomannan (Platelia Aspergillus enzyme immunoassay [EIA]; Bio-Rad Laboratories, Marnes-la-Coquette, France) monitoring was carried out. For each patient, age, sex, white blood cell count and neutrophil count at time of sampling, microbiology results related to the fever episode, underlying disease, antibiotic treatment, duration of antibiotic treatment after first sampling, chemotherapy, and, in case of stem cell transplantation, date of stem cell transplantation were retrieved from the patients' medical records. Patient characteristics are described in Table 1.
TABLE 1.
Patient characteristics (n = 74)
| Characteristic | Valuea |
|---|---|
| Age (mean [range]) (yr) | 52 (19–73) |
| Males/females | 45/29 |
| Underlying disease | |
| Hematologic malignancy | |
| Acute myeloid leukemia | 26 |
| Chronic myeloid leukemia | 2 |
| Myelodysplastic syndrome | 6 |
| Myeloproliferative neoplasm | 2 |
| Acute lymphoid leukemia | 4 |
| B-cell non-Hodgkin lymphoma | 23 |
| Hodgkin lymphoma | 1 |
| Combined T-cell/B-cell neoplasm | 1 |
| T-cell neoplasm | 5 |
| Nonhematologic malignancy | |
| Ewing sarcoma | 2 |
| Other | |
| Crohn's disease (stem cell transplantation) | 1 |
| Hemophagocytic syndrome | 1 |
| Blood culture bottles at start of fever (mean [range]) | 5 (2–8) |
| Patients with stem cell transplantation | |
| Allogeneic | 27 |
| Autologous | 19 |
| PCR/ESI-MS samples per patient (no. [range]) | 1 (1–4) |
| 1 | 54 |
| 2 | 10 |
| 3 | 9 |
| 4 | 1 |
| Absolute neutrophil count (109/liter) at time of sampling per episode | |
| <0.1 | 75 |
| 0.1–0.5 | 18 |
| 0.6–1.5 | 8 |
| >1.5 | 4 |
| Samples per febrile episode | |
| 1 | 95 |
| 2 | 5 |
| Days with intravenous antibiotic therapy after first sample per episode (mean [range]) | 12 (3–32) |
Values are the number, unless otherwise indicated.
Blood culture.
At the start of a febrile episode, at least two sets of blood cultures were collected (a set consists of one aerobic BacT/Alert FA bottle and one anaerobic FN FAN medium blood culture bottle [bioMérieux, Durham, NC, USA], each filled with 10 ml of blood). Blood cultures were collected from each lumen of the central venous catheter (CVC) and from a peripheral vein site. Blood specimens were processed by an automated blood culture system (BacT/Alert; bioMérieux) with 5 days of incubation. Identification of the bacteria and yeasts was performed by MALDI-TOF MS (Bruker Daltonics, Billerica, MA) and biochemical testing. Antimicrobial susceptibility testing (AST) was performed with Vitek 2 (bioMérieux, Marcy l'Etoile, France) or disk diffusion (Rosco Neo-Sensitabs, Taastrup, Denmark).
Iridica BAC BSI assay.
The 10-ml EDTA whole-blood sample was collected through the catheter at the same time of the blood cultures. Whole-blood samples were stored in their original EDTA tube at −20°C, as whole-blood samples, according to the instructions of the manufacturer. The Iridica BAC BSI assay (CE-IVD marked) was carried out, according to the instructions of the manufacturer. The different steps included sample preparation, PCR amplification, desalting of the PCR products, detection, and identification by ESI-TOF MS, as described previously (4). Briefly, after thawing, 5 ml of whole blood was chemically and mechanically lysed, and an extraction control was added to each specimen for process monitoring purposes. DNA extraction and PCR setup were automatically performed by a single instrument using prefilled individual disposable sample preparation cartridges and prefilled 16-well PCR strips. The BAC BSI assay utilizes several conserved-site primer pairs designed to amplify variable (and thereby discriminable) products from a broad range of bacteria and Candida spp., as well as primer pairs targeted to common antibiotic resistance loci conferring resistance to methicillin (mecA), vancomycin (vanA and vanB), and carbapenems (Klebsiella pneumoniae carbapenemase [KPC]). PCR products were then desalted and concentrated relative to human genomic DNA in an automated system and analyzed through ESI-MS. The base compositions of detected amplicon strands were deduced from the measured masses and compared with a reference database, leading to the identification of the microorganisms present in clinical samples. Internal calibrators present in each reaction allowed for a relative (qualitative) approximation of target concentrations (expressed as levels), which in turn were used to limit noise- and contamination-derived background detections through thresholding of positive signals. In order to detect possible contamination, a negative control provided by the manufacturer was analyzed in each extraction batch. After installation of the Iridica system, the Iridica DS/MS Check kit was run to control the performance of the desalter and mass spectrometer.
Analysis and definitions.
A BSI was defined by a positive blood culture and/or a positive PCR/ESI-MS sample. No clinical criteria were included in the evaluation of BSI, as in this population of neutropenic patients, signs and symptoms are nonspecific in a majority of the patients (fever, mucositis, and diarrhea), and in a majority of the cases, no other cultures are positive. One positive blood culture was considered diagnostic for a definite BSI, except when common skin contaminants were cultured (7), in which case at least two consecutive positive blood cultures drawn on separate occasions were needed for a definite BSI. BC collected from a central line and from a peripheral vein were also defined as separate occasions. If this was not the case, we needed to classify the case as a probable BSI if two BC collected from the same site were positive, or as no BSI if only one BC was positive. The following organisms fell into the category of common skin contaminants: coagulase-negative staphylococci, micrococci, Bacillus species (excluding Bacillus anthracis), Propionibacterium species, Corynebacterium species, viridans streptococci (excluding Streptococcus pneumoniae), and commensal Neisseria species. If organisms were reported as potential contaminants by the PCR/ESI-MS software, the BSI was also considered a probable BSI. For each case, the results obtained by PCR/ESI-MS and by blood culture were compared. All samples, including samples with detection of multiple microorganisms, were included in this comparison.
Clinical quantitative variables were compared between groups using the Mann-Whitney U test (nonnormal distribution), qualitative variables were compared by Pearson chi-square test, and data were expressed as the mean or median and/or range. Statistical analysis was performed using Analyse-it method validation edition software (Leeds, United Kingdom). P values of <0.05 were considered statistically significant.
RESULTS
In 74 consecutive patients, blood cultures and a single 10-ml EDTA whole-blood sample were collected at the start of each febrile neutropenic episode and, in case of persistent fever, also at day 5. A total of 106 blood samples from 100 different febrile episodes were available for PCR/ESI-MS testing. Data analysis was performed on 105 specimens, as in one case, an invalid PCR/ESI-MS result was obtained.
PCR/ESI-MS and blood culture results by specimen.
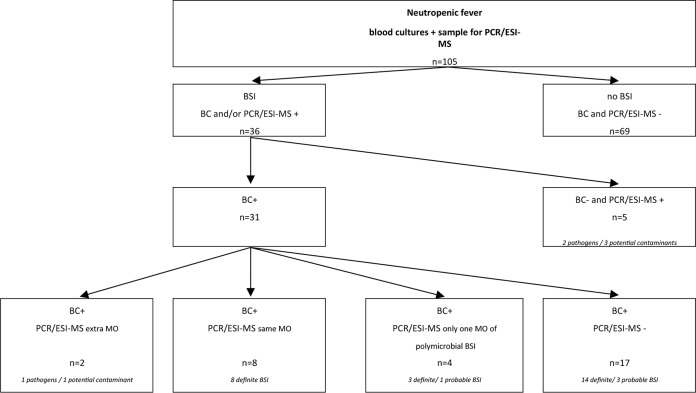
There was evidence of a bloodstream infection (BSI) in 36 cases (34%), with 14 cases having both PCR/ESI-MS and BC positivity, 17 cases having BC positivity only, and 5 cases having PCR/ESI-MS positivity only. In 69 cases, both PCR/ESI-MS and BC were negative. A polymicrobial BSI was diagnosed in 13 cases but detected by both methods in only one case. In 9 cases, a polymicrobial infection was detected by BC, but PCR/ESI-MS was negative (n = 4), only detected one microorganism (n = 4), or detected another microorganism (n = 1). In 3 cases, PCR/ESI-MS detected a polymicrobial infection with negative BC (n = 2) or only detected one microorganism by BC (n = 1).
The overall sensitivity of PCR/ESI-MS was 45% (14/31), specificity was 93% (69/74), and the negative predictive value was 80% (69/86) compared to blood cultures. Episodes with polymicrobial infection, where PCR/ESI-MS detected only the definite pathogen and not the potential contaminant(s), were categorized as true positive for PCR/ESI-MS compared to BC.
Table 2 summarizes all microorganisms detected by both methods, PCR/ESI-MS only, and BC only. Six Gram negatives and 22 Gram positives were detected by BC only. PCR/ESI-MS detected one Gram negative and 10 Gram positives not detected by conventional BC, while 17 microorganisms were detected by both methods.
TABLE 2.
Microorganisms detected by BC only, PCR/ESI-MS only, and detected by both methods for definite and probable BSI
| Organism reported | No. of microorganisms detected |
|||
|---|---|---|---|---|
| BC only | PCR/ESI-MS only | BC and PCR/ESI-MS | Total | |
| Definite BSI | ||||
| Gram negatives | 6 | 1 | 1 | 8 |
| Escherichia coli | 4 | 0 | 1 | 5 |
| Fusobacterium nucleatum | 0 | 1 | 0 | 1 |
| Klebsiella oxytoca | 1 | 0 | 0 | 1 |
| Klebsiella pneumoniae | 1 | 0 | 0 | 1 |
| Gram positives | 13 | 1 | 12 | 26 |
| Clostridium ramosum | 1 | 0 | 0 | 1 |
| Enterococcus faecium | 1 | 0 | 2 | 3 |
| Enterococcus faecalis | 1 | 0 | 1 | 2 |
| Gemella spp. | 2 | 0 | 0 | 2 |
| Rothia mucilaginosa | 1 | 0 | 2 | 3 |
| Staphylococcus epidermidisa | 4 | 0 | 5 | 9 |
| Staphylococcus haemolyticusa | 1 | 0 | 1 | 2 |
| Staphylococcus hominisa | 2 | 0 | 0 | 2 |
| Streptococcus mitis | 0 | 0 | 1b | 1 |
| Streptococcus pneumoniae | 0 | 1 | 0 | 1 |
| Fungi | 0 | 0 | 2 | 2 |
| Candida kefyr | 0 | 0 | 1c | 1 |
| Saccharomyces cerevisiae | 0 | 0 | 1c | 1 |
| Total | 19 | 2 | 15 | 36 |
| Probable BSI | ||||
| Brevibacterium paucivorans/Corynebacterium aurisa | 0 | 1 | 0 | 1 |
| Corynebacterium tuberculostearicuma | 0 | 1 | 0 | 1 |
| Gordonia polyisoprenivoransa | 0 | 2 | 0 | 2 |
| Micrococcus luteus | 1 | 0 | 0 | 1 |
| Propionibacterium acnesa | 0 | 2 | 0 | 2 |
| S. epidermidisa | 3 | 0 | 3 | 6 |
| S. haemolyticusa | 0 | 1 | 0 | 1 |
| S. hominisa | 4 | 0 | 0 | 4 |
| Total | 8 | 7 | 3 | 18 |
Reported as potential contaminant by PCR/ESI-MS BAC BSI assay.
PCR/ESI-MS reported Streptococcus pneumoniae.
PCR/ESI-MS reported “fungus detected, no ID provided.”
PCR/ESI-MS detected 3 definite BSI (Fusobacterium nucleatum [n = 1] and Streptococcus pneumoniae [n = 2]) and 5 probable BSI missed by BC (Fig. 1). The potential contaminants (Brevibacterium paucivorans/Corynebacterium auris, Propionibacterium acnes, Staphylococcus haemolyticus, Staphylococcus warneri/S. epidermidis/S. caprae together with detection of Corynebacterium tuberculostearicum, and Gordonia polyisoprenivorans) were detected in 4 patients with negative blood cultures and in 1 patient in whom another pathogen was detected by blood culture. A review of the patient records and other culture results was conducted for all patients. In one patient with S. haemolyticus definite BSI, both PCR/ESI-MS and BC detected the microorganism in the samples collected at start of fever. However, the sample collected at day 5 of the same episode was PCR/ESI-MS positive but culture negative. Bacterial DNA could still be detected after 4 days of vancomycin therapy in this patient, whereas blood cultures were already negative. In the two patients with S. pneumoniae detected by PCR/ESI-MS but not by BC, no evidence of pneumonia on chest radiography was found, and no other clinical specimens were positive. Interestingly, in one of these two patients with severe mucositis, all blood cultures (8/8 bottles) were positive with Streptococcus mitis, indicating that both PCR/ESI-MS and blood culture detected streptococci, but PCR/ESI-MS was potentially not able to differentiate between S. mitis and S. pneumoniae. The patient with Fusobacterium nucleatum detected by PCR/ESI-MS had severe mucositis and diarrhea during his conditioning for allogeneic stem cell transplantation. In the three patients with negative BC but for whom PCR/ESI-MS detected a definite BSI, patients were empirically treated with meropenem. This broad-spectrum antibiotic covered the detected microorganisms.
FIG 1.
Results of blood cultures (BC) and PCR/ESI-MS in the 105 cases of neutropenic fever. Bloodstream infection (BSI) included both definite and probable BSI based on BC and/or PCR/ESI-MS. MO, microorganism; +, positive result; −, negative result; definite BSI, at least one BC positive (except when a common skin contaminant is cultured, at least two consecutive blood cultures drawn on separate occasions are needed), or detection of a nonskin contaminant by PCR/ESI-MS; probable BSI, common skin contaminant cultured in two blood cultures drawn at the same site or detection of a potential contaminant by PCR/ESI-MS. BSI based on BC and/or PCR/ESI-MS.
On the other hand, in 17 cases with definite (n = 14) and probable (n = 3) BSI, PCR/ESI-MS was negative. Five of these patients had definite BSI with Gram-negative bacilli (Escherichia coli [n = 3] and Klebsiella sp. [n = 2]) and 9 patients with Gram-positive microorganisms.
Assessment of factors influencing performance of PCR/ESI-MS.
For the cases with definite BSI, based on positive BC (n = 27), the following variables and patient characteristics were compared between PCR/ESI-MS-nonconcordant (not detecting the same microorganism as in the blood culture) (n = 14) and PCR/ESI-MS-concordant (n = 13) samples: age, gender, white blood cell count, underlying malignancy, stem cell transplantation, chemotherapy and antibiotics at time of sampling, and number and sample site of (positive) BC bottles (Table 3). The cases with probable BSI based on blood culture (n = 4) were excluded in this comparison. In the cases with concordant results, the percentage of positive BC bottles was significantly higher than in the nonconcordant cases. No other significant associations were found.
TABLE 3.
Patient characteristics of patients having episodes with PCR/ESI-MS-discordant and -concordant results compared with BCa
| Patient characteristic | PCR/ESI-MS concordant (n = 13)b | PCR/ESI-MS nonconcordant (n = 14)c | Significance (P value) |
|---|---|---|---|
| Age (mean [range]) (yr) | 52 (19–65) | 50 (20–67) | 0.72 |
| Males/females | 8/5 | 7/7 | 0.36 |
| Patients with absolute WBC count (109/liter) at time of sampling of: | 0.24 | ||
| <0.5 | 11 | 13 | |
| 0.5–2 | 2 | 1 | |
| Patients with absolute neutrophil count (109/liter) at time of sampling of: | 0.57 | ||
| <0.1 | 11 | 13 | |
| 0.1–0.5 | 1 | 1 | |
| 0.6–1.5 | 1 | 0 | |
| >1.5 | 0 | 0 | |
| Blood culture bottles collected at time of PCR/ESI-MS sample (median [range]) | 8 (4–8) | 8 (2–8) | 0.61 |
| Percent blood culture bottles positive collected at the time of PCR/ESI-MS sample (mean [range]) | 78 (38–100) | 48 (13–100) | 0.02 |
| Patients with only positive blood culture(s) collected from a different site than the site where the PCR/ESI-MS sample was collected | 1 | 3 | |
| Patients with stem cell transplantation | 6 | 10 | 0.35 |
| Allogeneic | 5 | 7 | |
| Autologous | 1 | 3 | |
| Underling disease | |||
| Hematologic malignancy (%) | 13 (100) | 14 (100) | |
| Acute myeloid leukemia | 6 | 8 | |
| Chronic myeloid leukemia | 0 | 1 | |
| Myelodysplastic syndrome | 0 | 1 | |
| Myeloproliferative neoplasm | 0 | 0 | |
| Acute lymphoid leukemia | 4 | 0 | |
| B-cell non-Hodgkin lymphoma | 1 | 4 | |
| T-cell neoplasm | 2 | 0 | |
| Patients with chemotherapy at time of sampling (including 24 h before sample) | 5 | 4 | 0.55 |
| Patients (%) with antibiotic therapy at time of sampling using: | 13 (100) | 14 (100) | |
| Levofloxacin | 9 | 13 | |
| Meropenem | 2 | 1 | |
| Vancomycin | 1 | 0 | |
| Ceftazidime | 1 | 0 | |
| Fluconazole | 10 | 12 | |
| Acyclovir | 6 | 8 |
Values are the number, unless otherwise predicted.
PCR/ESI-MS and BC detected same MO.
BC positive and PCR/ESI-MS negative or MO not detected.
Detection of antibiotic resistance genes.
There were no identified cases of Klebsiella pneumoniae carbapenemase (KPC) or vancomycin-resistant enterococci, which was matched across PCR/ESI-MS and conventional AST. There were 5 detections of mecA coagulase-negative staphylococci, which was in agreement with the AST result.
Research analysis protocol with lower cutoff for detection.
PCR/ESI-MS results were reanalyzed by Abbott using a research analysis protocol with a lower cutoff for the detection for selected organisms, i.e., coagulase-negative Staphylococcus species, Lactobacillus species, and Aspergillus species, all of which have been previously reported to be isolated from symptomatic neutropenic patients. Propionibacterium acnes, Corynebacterium sp., Gordonia sp., Saccharomyces sp., nonpneumococcal Streptococcus sp. or, non-aeruginosa Pseudomonas species will not be reported if detected below the cutoff of the CE-marked protocol. Further, the updated protocol will also not distinguish between S. pneumoniae and S. mitis.
Using this modified protocol, three definite BSI were additionally detected compared to the standard Iridica protocol. Aspergillus fumigatus was detected in two samples from patients with negative blood cultures. At the time of sampling, these patients were diagnosed with probable invasive aspergillosis, according to the European Organization for Research and Treatment of Cancer (EORTC) criteria (8). The PCR/ESI-MS sample was collected at 2 days before and 2 days after daily serum galactomannan (GM) testing became positive (optical density index cutoff, 0.5). One other patient with probable invasive aspergillosis (IA) had a negative PCR/ESI-MS sample 1 day before GM became positive. In only one patient with definite BSI, the microorganism that was cultured from the blood but not detected with the standard Iridica analysis protocol could be detected by use of the lower cutoff. In this sample, Staphylococcus epidermidis was detected using the lower cutoff, in concordance with 7 of the 8 blood culture bottles collected at the same moment that were positive with S. epidermidis. Lowering the cutoff resulted in a sensitivity of 48% (15/31), a specificity of 91% (67/74), and a negative predictive value of 81% (67/83) compared to BC.
DISCUSSION
Our study in neutropenic patients demonstrates that PCR/ESI-MS, compared to standard BC, has an overall sensitivity of 47%, a specificity of 93%, and a negative predictive value of 81%. These results indicate a lower concordance between blood cultures and PCR/ESI-MS in the neutropenic population than that previously seen in other patient groups with normal white blood cell distribution (5, 6, 9–13), and a lower sensitivity than that of other PCR-based methods (5, 6, 9–13).
PCR/ESI-MS did detect in 9 patients additional pathogens compared to blood cultures but missed frequently encountered important pathogens. These results are not in accordance with a previous study comparing PCR/ESI-MS with blood culture in critically ill ICU patients (5). In that study, PCR/ESI-MS detected a pathogen in 37% of the cases compared to BC in only 11% of the cases. Possibly, the specific patient population of febrile neutropenic patients has influenced the sensitivity of PCR/ESI-MS. First, all patients had a low neutrophil count at the moment of sampling. Technically, the neutrophil and white blood cell counts in the sample can have a critical role in the extraction of the bacterial DNA. The lack of neutrophils containing microbial DNA might result in a lower concentration of microbial DNA in neutropenic patients. In our patient population, we did not find a correlation between the absolute white blood cell count or neutrophil count and the concordance of PCR/ESI-MS with BC results. However, all patients had low levels of white blood cells, resulting in only small differences in white blood cell count compared to the significantly higher white blood cell (WBC) counts in septic ICU or ER patients. A study including a control group with the same underlying conditions but with higher WBC counts is needed to clear out the role of the WBC count on the performance of PCR/ESI-MS. This study is also potentially informative to help design future studies of sensitive whole-blood technologies in cancer patients specifically. Second, in a majority of the patients, more than 4 blood culture bottles were collected at the time the PCR sample was drawn. From patients with a central venous catheter, each lumen of the catheter was sampled, in addition to a set of blood cultures collected peripherally. This resulted in a maximum of 8 bottles collected at the initiation of fever per patient. The positive effect of increasing the total blood volume on the sensitivity of blood cultures was demonstrated in several studies (14–16). Large volumes of blood culture may increase the risk of false-negative PCR/ESI-MS results performed on a single 5-ml EDTA sample. In our study, there was no difference regarding the number of BC bottles collected at the time of sampling between the PCR/ESI-MS-concordant and -discordant groups. However, in the group with PCR/ESI-MS-concordant results, a significantly higher percentage of BC bottles were positive than in the PCR/ESI-MS-discordant group, suggesting higher bacterial loads in the samples in the PCR/ESI-MS-concordant group. Third, the PCR/ESI-MS sample was collected through one lumen of the catheter, in contrast to the different sample sites for blood cultures (peripheral and through each lumen of the catheter). Some of the PCR/ESI-MS false-negative specimens were not collected from the same site as the positive blood culture draws, which potentially affected the results. Moreover, drawing the PCR/ESI-MS only from the catheter might reduce contamination, which may have introduced bias. To investigate the influence of the different sample sites, a prospective study should be set up in which the same sites are sampled for blood cultures and PCR. The antibacterial prophylaxis given to all study patients is another potentially influencing factor. This is in contrast to other patient populations for whom blood cultures are normally collected before starting antibiotics. Other potentially influencing factors were investigated by comparing patient characteristics between the PCR/ESI-MS-concordant and PCR/ESI-MS-discordant groups. No significant differences were detected in a comparison of underlying pathology, antibiotics or chemotherapy at the time of sampling, neutrophil count, age, and gender between the two groups.
Lowering the cutoff for detection in the reanalysis and expanding the reportable organisms to include Aspergillus species led to the detection of A. fumigatus, which was not detected using the CE-marked protocol. A. fumigatus detection in these two patients was in concordance with the diagnosis of probable invasive fungal infection. On the other hand, lowering the cutoff could have the unintended consequence of detection of reagent contamination, as described previously for A. fumigatus (17), or increased detection of potential contaminants, such as P. acnes. The protocol evaluated here was selective to avoid environmental organisms from being overreported. Further optimization of the detection cutoff could possibly increase sensitivity in the neutropenic patient population.
The lower sensitivity of PCR/ESI-MS in this population is inconsistent with the results of other molecular approaches in the same patient population of neutropenic patients. During the last 5 years, different molecular tests were studied in the febrile neutropenic population, including SeptiFast (Roche Diagnostics GmbH, Manheim, Germany), SepsiTest (UMD, Molzym, Bremen, Germany), and DNA microarray hybridization (9–12, 18–20). Compared to blood cultures, sensitivities were reported ranging from 46% in pediatric hematology-oncology patients to 91% in adult neutropenic patients (10, 18). Similar to PCR/ESI-MS, although less frequently, these PCR-based methods also failed in some cases to detect Gram negatives and Gram positives, but the detection of additional microorganisms (including fungi and fastidious microorganisms) and clinically relevant microorganisms in BC-negative patients during antibiotic therapy, and the faster results were seen as advantages compared to conventional BC. Recently, Idelevich et al. conducted a randomized controlled study to investigate the impact of multiplex PCR on antimicrobial treatment in febrile patients (21). The additional multiplex PCR testing to conventional BC accelerated the switch to targeted antimicrobial therapy (median time, 21.4 h) compared to that of the control group (median time, 47.5 h), but no significant differences were seen in patient outcomes.
Another limitation of PCR/ESI-MS is the very limited information on antimicrobial susceptibility, unlike culture techniques, hampering its use in clinical practice to guide antimicrobial therapy, especially in a setting in which multiresistant microorganisms can be expected. Expansion of the available molecular targets (mecA, KPC, vanA, and vanB) to other resistance mechanisms is warranted. We hypothesize that the clinical impact of the additional identifications by PCR/ESI-MS in our study population would be small. The association of vancomycin with the empirically started meropenem therapy could be needed in case coagulase-negative staphylococci are detected. However, in the other cases, the detected microorganisms were probably covered by meropenem, and the absence of susceptibility results would hamper the clinician from suspending the meropenem and switching to a narrower-spectrum antibiotic.
Besides these limitations, the advantages of the BAC BSI assay compared to other molecular PCR-based assays and conventional blood culture are its short turnaround time (less than 8 h), the nearly totally automated platform, and the possibility of detecting more than 750 different bacterial and Candida species. However, the high cost of the equipment and reagents compared to blood cultures, the big footprint of the system, the need for a trained technician to perform the few manual steps, by preference one who is available at night and/or during the weekend, will have to be taken into account to evaluate the potential of the assay in clinical practice.
Finally, we aimed to evaluate the performance of PCR/ESI-MS in comparison with blood culture in neutropenic patients. Clearly, future studies, preferably multicentric ones including larger numbers of patients and relevant control groups, are needed to confirm our results and to better understand the underlying factors leading to the lower sensitivity of PCR/ESI-MS in this particular patient population. Nowadays, broad-spectrum antibiotics are often given until neutrophil recovery, even in patients with noninfectious fever. Designing a tool that can discriminate between infectious and noninfectious causes, allowing clinicians to withhold or early withdraw unnecessary antibacterial therapy, and that at the same time can detect more clinically relevant pathogens than standard blood cultures, enabling early targeted therapy, is the ultimate goal. Such tools can then be incorporated and investigated in neutropenic care pathways, comparing the outcome of a standard empirical approach to an approach based upon their implementation (a preemptive approach).
To conclude, compared to blood cultures, PCR/ESI-MS testing detected additional microorganisms in 7 cases (7% of all samples [n = 105] collected during a febrile episode). The negative predictive value of PCR/ESI-MS was only 82%; as such, a negative result cannot be relied on to rule out a BSI or to stop antibiotics in neutropenic patients. Our study reports a lower concordance between BC and PCR/ESI-MS than previously seen in other patient groups and a lower sensitivity than other PCR-based methods.
ACKNOWLEDGMENTS
We thank Rangarajan Sampath (Ibis Biosciences, Abbott) for critically reviewing the manuscript.
K. Lagrou has received research grants, travel support, and lecture honoraria from Gilead, MSD, and Pfizer. J. Maertens has served as consultant to Schering-Plough, Gilead Sciences, Merck, Sharp & Dohme, Pfizer, Inc., Bio-Rad, Fujisawa Health Care Inc., Astellas, Nextar, Zeneus (Cephalon), ViroPharma, and Boehringer-Ingelheim; has received research funding from Bio-Rad, Merck, Sharp & Dohme, and Pfizer Inc.; and is on the speaker's bureau for Schering-Plough, Gilead Sciences, Merck, Sharp & Dohme, Pfizer, Inc., Bio-Rad, Fujisawa Health Care, Inc., Astellas, and Zeneus (Cephalon).
Ibis Biosciences, Abbott provided the PCR/electrospray ionization-mass spectrometry devices and reagents in the framework of an early access program.
REFERENCES
- 1.Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, Raad II, Rolston KV, Young JA, Wingard JR, Infectious Diseases Society of America. 2011. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 52:e56–e93. doi: 10.1093/cid/cir073. [DOI] [PubMed] [Google Scholar]
- 2.Klastersky J, Ameye L, Maertens J, Georgala A, Muanza F, Aoun M, Ferrant A, Rapoport B, Rolston K, Paesmans M. 2007. Bacteraemia in febrile neutropenic cancer patients. Int J Antimicrob Agents 30(Suppl 1):S51–S59. [DOI] [PubMed] [Google Scholar]
- 3.Ecker DJ, Sampath R, Li H, Massire C, Matthews HE, Toleno D, Hall TA, Blyn LB, Eshoo MW, Ranken R, Hofstadler SA, Tang YW. 2010. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev Mol Diagn 10:399–415. doi: 10.1586/erm.10.24. [DOI] [PubMed] [Google Scholar]
- 4.Bacconi A, Richmond GS, Baroldi MA, Laffler TG, Blyn LB, Carolan HE, Frinder MR, Toleno DM, Metzgar D, Gutierrez JR, Massire C, Rounds M, Kennel NJ, Rothman RE, Peterson S, Carroll KC, Wakefield T, Ecker DJ, Sampath R. 2014. Improved sensitivity for molecular detection of bacterial and Candida infections in blood. J Clin Microbiol 52:3164–3174. doi: 10.1128/JCM.00801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent JL, Brealey D, Libert N, Abidi NE, O'Dwyer M, Zacharowski K, Mikaszewska-Sokolewicz M, Schrenzel J, Simon F, Wilks M, Picard-Maureau M, Chalfin DB, Ecker DJ, Sampath R, Singer M, Rapid Diagnosis of Infections in the Critically Ill Team. 2015. Rapid diagnosis of infection in the critically ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med 43:2283–2291. doi: 10.1097/CCM.0000000000001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordana-Lluch E, Giménez M, Quesada MD, Rivaya B, Marcó C, Domínguez MJ, Arméstar F, Martró E, Ausina V. 2015. Evaluation of the broad-range PCR/ESI-MS technology in blood specimens for the molecular diagnosis of bloodstream infections. PLoS One 10:e0140865. doi: 10.1371/journal.pone.0140865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chizuka A, Kami M, Kanda Y, Murashige N, Kishi Y, Hamaki T, Kim SW, Hori A, Kojima R, Mori SI, Tanosaki R, Gomi H, Takaue Y. 2005. Value of surveillance blood culture for early diagnosis of occult bacteremia in patients on corticosteroid therapy following allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant 35:577–582. doi: 10.1038/sj.bmt.1704830. [DOI] [PubMed] [Google Scholar]
- 8.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bravo D, Blanquer J, Tormo M, Aguilar G, Borras R, Solano C, Clari MA, Costa E, Munoz-Cobo B, Argueso M, Pineda JR, Navarro D. 2011. Diagnostic accuracy and potential clinical value of the LightCycler SeptiFast assay in the management of bloodstream infections occurring in neutropenic and critically ill patients. Int J Infect Dis 15:e326–331. doi: 10.1016/j.ijid.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Guido M, Quattrocchi M, Zizza A, Pasanisi G, Pavone V, Lobreglio G, Gabutti G, De Donno A. 2012. Molecular approaches in the diagnosis of sepsis in neutropenic patients with haematological malignances. J Prev Med Hyg 53:104–108. [PubMed] [Google Scholar]
- 11.Lamoth F, Jaton K, Prod'hom G, Senn L, Bille J, Calandra T, Marchetti O. 2010. Multiplex blood PCR in combination with blood cultures for improvement of microbiological documentation of infection in febrile neutropenia. J Clin Microbiol 48:3510–3516. doi: 10.1128/JCM.00147-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paolucci M, Stanzani M, Melchionda F, Tolomelli G, Castellani G, Landini MP, Varani S, Lewis RE, Sambri V. 2013. Routine use of a real-time polymerase chain reaction method for detection of bloodstream infections in neutropaenic patients. Diagn Microbiol Infect Dis 75:130–134. doi: 10.1016/j.diagmicrobio.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 13.von Lilienfeld-Toal M, Lehmann LE, Raadts AD, Hahn-Ast C, Orlopp KS, Marklein G, Purr I, Cook G, Hoeft A, Glasmacher A, Stüber F. 2009. Utility of a commercially available multiplex real-time PCR assay to detect bacterial and fungal pathogens in febrile neutropenia. J Clin Microbiol 47:2405–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall MM, Ilstrup DM, Washington JA Jr. 1976. Effect of volume of blood cultured on detection of bacteremia. J Clin Microbiol 3:643–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenney JH, Reller LB, Mirrett S, Wang WL, Weinstein MP. 1982. Controlled evaluation of the volume of blood cultured in detection of bacteremia and fungemia. J Clin Microbiol 15:558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cockerill FR III, Wilson JW, Vetter EA, Goodman KM, Torgerson CA, Harmsen WS, Schleck CD, Ilstrup DM, Washington JA Jr, Wilson WR. 2004. Optimal testing parameters for blood cultures. Clin Infect Dis 38:1724–1730. doi: 10.1086/421087. [DOI] [PubMed] [Google Scholar]
- 17.Harrison E, Stalhberger T, Whelan R, Sugrue M, Wingard JR, Alexander BD, Follett SA, Bowyer P, Denning DW, Aspergillus Technology Consortium (AsTeC). 2010. Aspergillus DNA contamination in blood collection tubes. Diagn Microbiol Infect Dis 67:392–394. doi: 10.1016/j.diagmicrobio.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shachor-Meyouhas Y, Sprecher H, Moscoviz D, Zaidman I, Haimi M, Kassis I. 2013. Molecular-based diagnosis of bacteremia in the setting of fever with or without neutropenia in pediatric hematology-oncology patients. J Pediatr Hematol Oncol 35:500–503. doi: 10.1097/MPH.0b013e31829eec78. [DOI] [PubMed] [Google Scholar]
- 19.Negoro E, Iwasaki H, Tai K, Ikegaya S, Takagi K, Kishi S, Yamauchi T, Yoshida A, Urasaki Y, Shimadzu M, Ueda T. 2013. Utility of PCR amplification and DNA microarray hybridization of 16S rDNA for rapid diagnosis of bacteremia associated with hematological diseases. Int J Infect Dis 17:e271–e276. doi: 10.1016/j.ijid.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Wellinghausen N, Kochem AJ, Disque C, Muhl H, Gebert S, Winter J, Matten J, Sakka SG. 2009. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J Clin Microbiol 47:2759–2765. doi: 10.1128/JCM.00567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Idelevich EA, Silling G, Niederbracht Y, Penner H, Sauerland MC, Tafelski S, Nachtigall I, Berdel WE, Peters G, Becker K, Molecular Diagnostics of Sepsis Study Group. 2015. Impact of multiplex PCR on antimicrobial treatment in febrile neutropenia: a randomized controlled study. Med Microbiol Immunol 204:585–592. doi: 10.1007/s00430-014-0385-7. [DOI] [PubMed] [Google Scholar]