Abstract
Bovine anaplasmosis caused by the intraerythrocytic rickettsial pathogen Anaplasma marginale is endemic in South Africa. Anaplasma marginale subspecies centrale also infects cattle; however, it causes a milder form of anaplasmosis and is used as a live vaccine against A. marginale. There has been less interest in the epidemiology of A. marginale subsp. centrale, and, as a result, there are few reports detecting natural infections of this organism. When detected in cattle, it is often assumed that it is due to vaccination, and in most cases, it is reported as coinfection with A. marginale without characterization of the strain. A total of 380 blood samples from wild ruminant species and cattle collected from biobanks, national parks, and other regions of South Africa were used in duplex real-time PCR assays to simultaneously detect A. marginale and A. marginale subsp. centrale. PCR results indicated high occurrence of A. marginale subsp. centrale infections, ranging from 25 to 100% in national parks. Samples positive for A. marginale subsp. centrale were further characterized using the msp1aS gene, a homolog of msp1α of A. marginale, which contains repeats at the 5′ ends that are useful for genotyping strains. A total of 47 Msp1aS repeats were identified, which corresponded to 32 A. marginale subsp. centrale genotypes detected in cattle, buffalo, and wildebeest. RepeatAnalyzer was used to examine strain diversity. Our results demonstrate a diversity of A. marginale subsp. centrale strains from cattle and wildlife hosts from South Africa and indicate the utility of msp1aS as a genotypic marker for A. marginale subsp. centrale strain diversity.
INTRODUCTION
Bovine anaplasmosis (gallsickness) is a tick-borne disease caused by the intraerythrocytic rickettsial pathogen Anaplasma marginale (1). A. marginale is globally prevalent and results in anemia, with mortality rates of up to 30% (2). Anaplasma marginale subspecies centrale is a less virulent subspecies detected by Sir Arnold Theiler, who recognized its potential as a vaccine against anaplasmosis; 100 years later this live vaccine is still in use in South Africa, Israel, South America, and Australia (3, 4). The strain that is used as a vaccine originated from Theiler's original isolation and was exported at various times to other countries where it has been propagated in the laboratory; the strain known as the “Israel strain” or the “vaccine strain” was sent to Israel in the 1950s and was used to generate the complete genome sequence for A. marginale subsp. centrale in 2010 (5). A. marginale subsp. centrale does not provide complete protection against A. marginale infection but does protect against severe anaplasmosis (6, 7).
A. marginale infects a wide range of ruminants including buffalo (Bubalus bubalis and Syncerus caffer), wildebeest (Connochaetes gnou and Connochaetes taurinus), American bison (Bison bison), white-tailed deer (Odocoileus virginianus), mule deer (Odocoileus hemionus hemionus), black-tailed deer (Odocoileus hemionus columbianus), and Rocky Mountain elk (Cervus elaphus nelsoni) (8–11). Cattle are naturally susceptible to A. marginale (4). There has not been much interest in the epidemiology of A. marginale subsp. centrale, with few reports detecting natural infections of this organism; most often, when detected in cattle it is assumed that it is due to vaccination and is reported as coinfection with A. marginale without characterization of the strain (12). Reported A. marginale subsp. centrale single infections were detected by the reverse line blot (RLB) hybridization assay in Italy without characterization of the strain. More recently, the first known case of bovine anaplasmosis caused by A. marginale subsp. centrale in Europe was reported (13). While this study described genetic heterogeneity of A. marginale subsp. centrale strains from different geographic areas in Italy, it is not clear how these are related to the vaccine strain.
For A. marginale, the Msp1a protein/gene (msp1α) has been used as a genotypic marker to differentiate strains (14). Msp1a is encoded by the single-copy gene, msp1α and differs among strains due to variable sequence and numbers of an 84/87-bp repeat sequence (28 or 29 amino acids) located near the amino terminus of the protein (14). A number of studies have examined Msp1a repeats in the United States, South America, Australia, the Philippines, Europe, Israel, China, and Mexico, resulting in identification of more than 200 repeats (14–16). In South Africa, two studies have been conducted to genetically characterize strains using msp1α (17, 18), revealing that the repeat structure is common between South African, American, and European strains of A. marginale; in fact, some of the repeat sequences that were detected were identical to ones that were detected in the United States. Not surprisingly, there were also new repeat sequences detected that are, thus far, unique to South Africa.
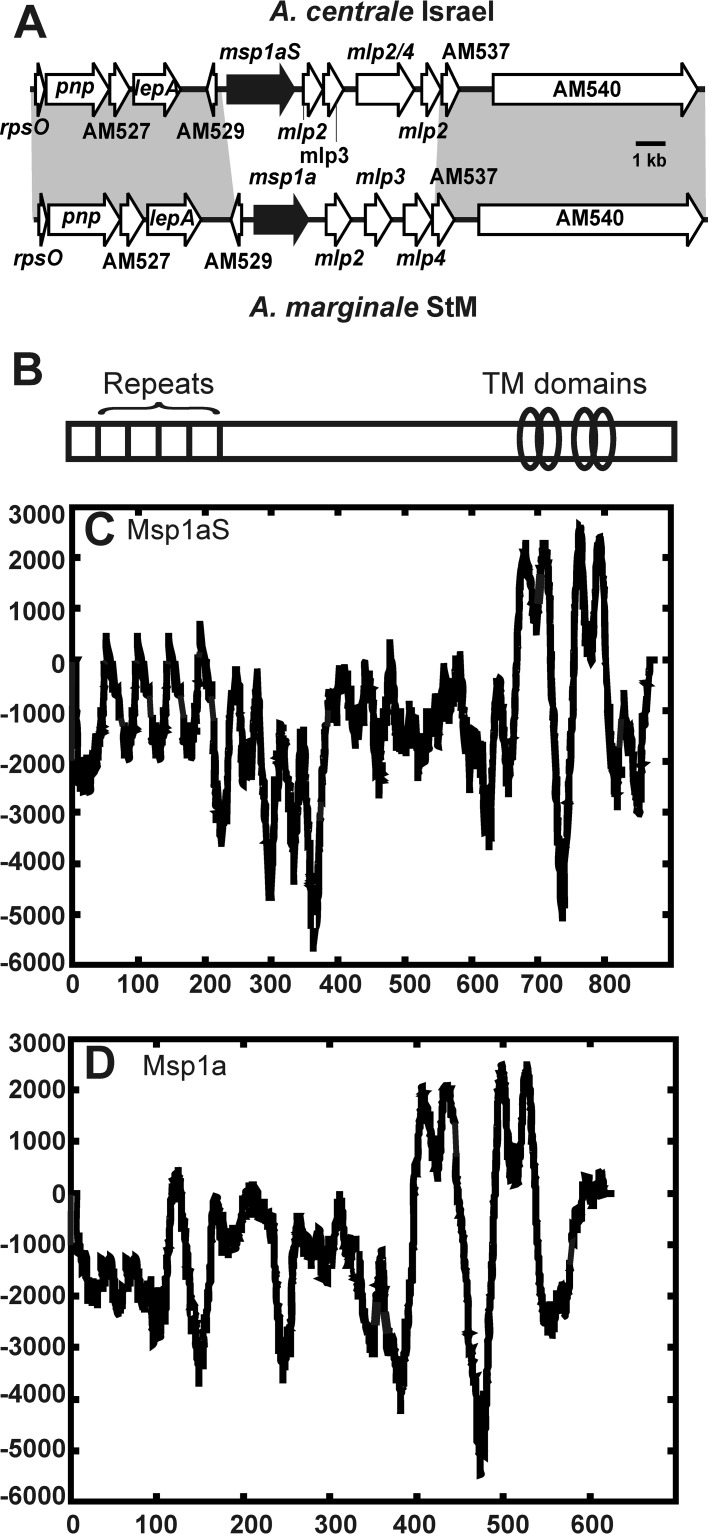
A. marginale subsp. centrale was thought not to have a homolog of msp1α; however, complete genome sequencing of the Israel vaccine strain revealed that there is a gene that resides in a position syntenic to A. marginale msp1α (5). This gene was named msp1aS (S for syntenic; a gene flanked by the same set of genes in two genomes) and has 31 to 36% amino acid sequence identity depending on the A. marginale strain compared. Importantly, there are structural similarities, including repeats near the amino terminus and two sets of transmembrane domains near the carboxy terminus that indicate that these proteins are likely homologs (Fig. 1). The repeats in A. marginale subsp. centrale strain Israel Msp1aS are longer (47 amino acids in length) than the A. marginale Msp1a repeats, and there is no sequence identity between the repeats in the two organisms. The vaccine strain has four repeats with an msp1aS genotype of Ac1 Ac1 Ac1 Ac2.
FIG 1.
Schematic representation and TMpred plots of Msp1aS. (A) Genomic positioning of Msp1aS of A. marginale subsp. centrale, also showing that it is syntenic to Msp1a of A. marginale St. Maries strain (StM), which suggests that these proteins are homologs. (B) While there is little sequence conservation, these proteins have similar structures: both have a set of repeats near the amino terminus and two sets of transmembrane (TM) domains toward the carboxy terminus. (C and D) TMpred plots show the transmembrane prediction profile for both molecules (Msp1aS from the fully sequenced Israel strain of A. marginale subsp. centrale and Msp1a from the fully sequenced St. Maries strain of A. marginale). Values greater than 500 (y axis) indicate transmembrane domains. The repeats of Msp1aS are almost twice as long as those of Msp1a.
In the present study, we have used a duplex quantitative real-time PCR (qPCR) assay to screen for the presence of A. marginale subsp. centrale and A. marginale in vaccinated and unvaccinated cattle and wildlife, indicating that these infections are common and often occur as mixed infections. Samples that tested positive using this screen were then further analyzed for the msp1aS genotype, demonstrating that the vaccine strain genotype is prevalent in cattle herds that practice vaccination, while other more divergent genotypes are present in wildlife species.
MATERIALS AND METHODS
Blood collection and DNA extraction.
A total of 380 blood samples from wild ruminant species including African buffalo (Syncerus caffer, n = 97); waterbuck (Kobus ellipsiprymnus, n = 14); eland (Taurotragus oryx, n = 23); black wildebeest (Connochaetes gnou, n = 54); and blue wildebeest (Connochaetes taurinus, n = 23), together with 86 cattle samples, were obtained from the Wildlife Biological Resource Center (WBRC) and Biobank South Africa (SA) under the auspices of the National Zoological Gardens of South Africa (NZG) and from the South African National Parks (SANParks) Biobank. The remaining buffalo blood samples (n = 41) were made available to us by Dave Cooper from Hluhluwe-iMfolozi Park. Additionally, 42 blood samples from vaccinated cattle were obtained from two commercial farms in Bergville, KwaZulu-Natal, South Africa (Table 1). Standard techniques were followed in collecting blood samples for laboratory examination. Genomic DNA was extracted using the QIAmp DNA extraction kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. DNA was eluted in 100 μl of elution buffer and stored at −20°C.
TABLE 1.
Host samples used in this study
| Sample no. | Species | No. of samples | Sample type | Collection site | Origina | Province |
|---|---|---|---|---|---|---|
| 565–614 | Buffalo | 50 | EDTA-blood | SANParksb | KNP | Mpumalanga |
| 974–987 | Buffalo | 14 | EDTA-blood | SANParks | CNP | Eastern Cape |
| 1002–1016 | Buffalo | 15 | EDTA-blood | SANParks | AEP | Eastern Cape |
| 988–995 and 66/13 | Buffalo | 9 | EDTA-blood | SANParks | GNP | Northern Cape |
| 998–1001 and 1017–1021 | Buffalo | 9 | EDTA-blood | SANParks | MNP | Northern Cape |
| 1–41 | Buffalo | 41 | EDTA-blood | HiP | HiP | KwaZulu-Natal |
| 924–937 and 947–955 | Black wildebeest | 23 | EDTA-blood | SANParks | MTNZNP | Eastern Cape |
| 938–939 | Black wildebeest | 2 | EDTA-blood | SANParks | TMNP | Western Cape |
| 942, 944–953 and 955–972 | Black wildebeest | 29 | EDTA-blood | SANParks | MNP | Northern Cape |
| 1036–1056 | Blue wildebeest | 21 | EDTA-blood | SANParks | MNP | Northern Cape |
| 1057–1058 | Blue wildebeest | 2 | EDTA-blood | SANParks | WCNP | Western Cape |
| 1022–1031 | Eland | 10 | EDTA-blood | SANParks | MNP | Northern Cape |
| 1032–1035 | Eland | 4 | EDTA-blood | SANParks | AEP | Eastern Cape |
| 459–467 | Eland | 9 | FTA filter paper | WBRC, NZGc | NZG | Gauteng |
| 1059–1062 | Waterbuck | 4 | EDTA-blood | SANParks | MNP | Northern Cape |
| 468–470 | Waterbuck | 3 | FTA filter paper | WBRC, SA, NZG | Rietvlei NR, JHB Zoological Gardens, Mohale Gate (Gauteng area) | Gauteng |
| 543, 549 | Waterbuck | 2 | EDTA-blood | WBRC, SA, NZG | KNP | Mpumalanga |
| 544–548 | Waterbuck | 5 | EDTA-blood | WBRC, SA, NZG | MaNP | Limpopo |
| WC103–WC128 | Cattle | 26 | EDTA-blood | NZG collection | WC F3d | Western Cape |
| KZN129–KZN158 | Cattle | 30 | EDTA-blood | NZG collection | KZN F4 | KwaZulu-Natal |
| FS1–FS30 | Cattle | 30 | EDTA-blood | NZG collection | FS F5 | Free State |
| Berg 1–Berg 21 | Cattle | 21 | EDTA-blood | Bergville farm | Bergville F1 | KwaZulu-Natal |
| Berg 22–Berg 42 | Cattle | 21 | EDTA-blood | Bergville farm | Bergville F2 | KwaZulu-Natal |
Origin, the park/farm from where the sample originates: Kruger National Park (KNP), Cambedoo National Park (CNP), Graspan National Park (GNP), Mokala National Park (MNP) Addo Elephant Park (AEP), Hluhluwe-iMfolozi Park (HiP), Mountain Zebra National Park (MTNZNP), Table Mountain National Park (TMNP), West Coast National Park (WCNP), Marakele National Park (MaNP).
SANParks, South African National Parks.
WBRC, Wildlife Biological Research Center; NZG, National Zoological Gardens, South Africa.
F, farm.
The study was approved by the Animal Ethics Committee of the University of Pretoria, South Africa (V085-14), and permission to use wildlife samples was given by SANParks Biobank under reference number LARBJ1118 Conservation Genetics, by the WBRC, and by Biobank SA under the auspices of the NZG of South Africa and the Johannesburg Zoo with project number NZG/P13/05. Collection of cattle samples was approved by the Department of Agriculture, Forestry and Fisheries under section 20 of the Animal Diseases Act of 1984 with reference 12/11/1/1.
Duplex real-time PCR assay.
Quantitative real-time PCR (qPCR) for simultaneous detection and quantification of A. marginale and A. marginale subsp. centrale DNA was performed as described previously (19) with some modifications for use on a LightCycler real-time machine (28) (Roche Diagnostics, Mannheim, Germany). The qPCR was performed in a final reaction volume of 20 μl, containing 2 μl of DNA template (100 to 200 ng of DNA), 12.5 μl of FastStart DNA Master hybridization mix (Roche Diagnostics, Mannheim, Germany), 600 nmol/liter of A. marginale-specific primers AM-For (5′ TTG GCA AGG CAG CAG CTT 3′) and AM-Rev (5′-TTC CGC GAG CAT GTG CAT-3′), 900 nmol/liter of A. marginale subsp. centrale-specific primers AC-For (5′-CTA TAC ACG CTT GCA TCT C-3′) and AC-Rev (5′-CGC TTT ATG ATG TTG ATG C-3′), and 200 nmol/liter of probes AM-Pb (5′-6FAM-TCG GTC TTA ACA TCT CCA GGC TTT CAT-BHQ1-3′) and AC-Pb (5′-LC610-ATC ATC ATT CTT CCC CTT TAC CTC GT-BHQ2-3′). Thermal cycling conditions were as follows: UDG activation at 40°C for 10 min, preincubation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 1 min and annealing-extension at 60°C for 1 min, and a final cooling step at 40°C for 30 s. The results were analyzed using LightCycler software version 4.0 (Roche Diagnostics). The software indicates a positive result by a Cq value (quantification cycle, synonymous with the Cp, crossing point, value given by the LightCycler instrument), at which fluorescence from amplification exceeds the background fluorescence, and a score of 1 to 5. Negative samples have a score of −1 to −5 and no Cp values. A lower Cq correlates with a higher starting concentration of target DNA in a sample, which then indicates a positive infection. FAM fluorescence (530 nm) was generated in A. marginale-positive samples and LC-610 (610 nm) signals were generated in A. marginale subsp. centrale-positive samples. DNA extracted from the A. marginale subsp. centrale vaccine strain (Onderstepoort Biological Products [OBP], Pretoria, South Africa) was used as a positive control, and samples C14, C57, or F48 (originating from cattle in the Mnisi Community area, Mpumalanga, South Africa) were used as positive controls for A. marginale. The presence of A. marginale in these samples was confirmed by sequencing of the msp1β genes. A negative and a positive control were included in each set of PCRs that was performed. The analytical specificity of the assay was determined by analyzing DNA from closely related species such as Anaplasma sp. Omatjenne and A. phagocytophilum (20). The efficiency of the assay was determined from 10-fold serial dilutions of plasmid DNA from clones 9410c (A. marginale subsp. centrale) and F48a (A. marginale).
Analysis of the msp1aS gene.
A. marginale subsp. centrale-positive samples which had low Cq values as detected by qPCR were selected for analysis of the msp1aS gene. Primers MSP1asFZ (5′-CAA GGT CAA GAG TCA GCA TCA TCA GAT G-3′) and MSP1asRZ (5′-CTC CGC GCA CAA TAC TTT CAA CCT CC-3′) were designed based on the A. marginale subsp. centrale genome sequence (GenBank accession CP001759) to target tandem repeats within the msp1aS gene. PCR was performed in a final reaction volume of 25 μl containing Phusion Flash high-fidelity PCR master mix (Thermo Fisher Scientific), 10 pM of each primer, and genomic DNA. Thermal cycling was carried out in a Veriti thermal cycler (Thermo Fisher Scientific) and consisted of an initial denaturation at 98°C for 10 s, followed by 30 cycles of denaturation at 98°C for 1 s, annealing at 67°C for 30 s and extension at 72°C for 15 s, and a final extension at 72°C for 1 min. DNA extracted from the A. marginale subsp. centrale vaccine obtained from OBP (Pretoria, South Africa) was used as a positive control.
Purified PCR amplicons were cloned into the pJET vector (Thermo Fisher Scientific). Recombinant plasmids were isolated using a High Pure plasmid isolation kit (Roche Diagnostics, Mannheim, Germany) and sequenced using 1 μl of 2 μM M13 primers with an ABI BigDye v3.1 kit on an ABI 3500xL genetic analyzer at Inqaba Biotec (Pretoria, South Africa).
Sequences were assembled, edited, and translated to amino acids using CLC Main Workbench 7.0.3 (Qiagen, Denmark). Tandem repeats were identified using Tandem Repeats Finder (https://tandem.bu.edu/trf/trf.html) (21). The repeats were named Acn, to distinguish them from A. marginale Msp1a repeats. Truncated repeats were designated with a T at the end of the name. Repeats were curated and analyzed using RepeatAnalyzer (29). Repeat sequences were aligned using the AlignX module of Vector NTI (Invitrogen).
Diversity measures.
RepeatAnalyzer calculates four genetic diversity metrics, each of which captures the diversity of repeats in a geographic region in a different way. Broadly, they fall into two groups, those that measure the amount of different repeats and those that measure the distribution of those repeats. Within each of these categories there is a global and a local formulation. The local version of a metric calculates the score independently on each genotype and averages these together to get the final score, while the global version looks at all genotypes together. Specifically, the GDM1-L score can be interpreted as the percentage of unique repeats in each genotype in the region, while the GDM1-G score is the percentage of unique repeats across all genotypes in the region. The GDM2-L score can be interpreted as the amount of variation (measured as standard deviation) in the number of occurrences of the repeats in a genotype, while the GDM2-G score is the amount of variation in the number of occurrences of all the repeats in all genotypes in the region. A high GDM1 score means that there are more unique repeats, with 0 as the minimum (when all repeats are the same) and 1 being the maximum (when each repeat is unique). A high GDM2 score means that the repeats are distributed more unevenly, with a minimum of 0 (when all repeats occur the same number of times) and values ranging up to but not including 0.5 as the unevenness of repeat distribution increases.
RESULTS
Occurrence of Anaplasma species in wild ruminants and cattle in South Africa.
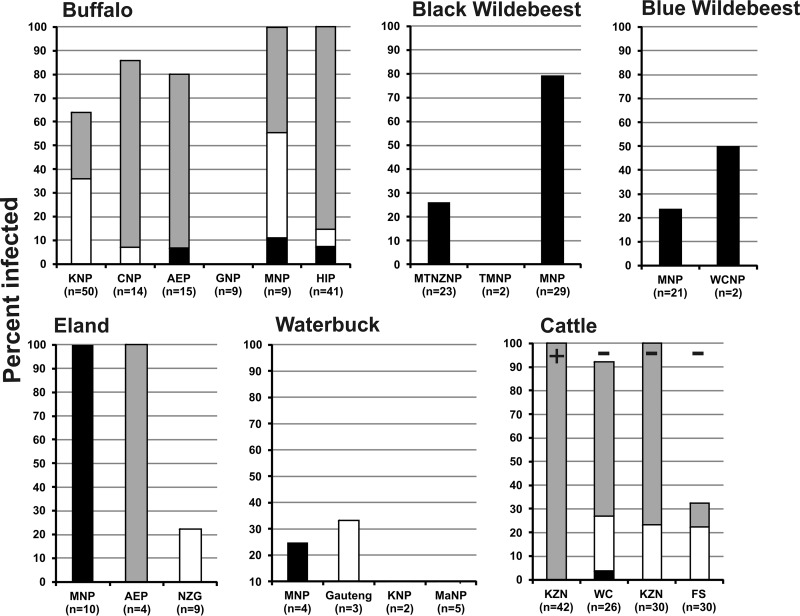
Duplex qPCR results indicated that A. marginale subsp. centrale single infections are common among black wildebeest (Mokala National Park [MNP], 79.3%), blue wildebeest (West Coast National Park [WCNP], 50%), waterbuck (MNP, 25%), and eland (MNP, 100%). Wildebeest did not harbor any A. marginale infections. Mixed infections were frequently found in both buffalo and cattle, ranging from 28% to 100% of animals from a given area being positive for both A. marginale and A. marginale subsp. centrale infections. Buffalo samples had high rates of mixed infections and also had lower rates of single infections with A. marginale subsp. centrale than with A. marginale. Interestingly, single infections of both species predominated in sets of animals from specific parks (see eland and waterbuck in Fig. 2), indicating that environment plays a role in exposure to the two pathogens.
FIG 2.
Stacked bar graphs showing occurrence of Anaplasma species in wild ruminants and cattle. Buffalo, black and blue wildebeest, eland, waterbuck, and cattle were analyzed by duplex real-time PCR. Animals were sampled from the following national parks and provinces: Kruger National Park (KNP), Cambedoo National Park (CNP), Addo Elephant Park (AEP), Graspan National Park (GNP), Mokala National Park (MNP), Hluhluwe-iMfolozi Park (HIP), Mountain Zebra National Park (MTNZNP), Table Mountain National Park (TMNP), West Coast National Park (WCNP), National Zoological Gardens of South Africa (NZG), Marakele National Park (MaNP), KwaZulu-Natal (KZN), Western Cape (WC), and Free State (FS). Numbers in parentheses indicate the total numbers of animals sampled from that park/province. Samples were collected from vaccinated (+) and unvaccinated (−) cattle. Black indicates animals positive for A. marginale subsp. centrale, gray indicates animals with mixed infections, and white indicates animals positive for A. marginale.
Characterization of MSP1aS.
Because the sequenced Israel vaccine strain was removed from South Africa more than 60 years ago, we obtained a batch of the vaccine currently produced at OBP in Pretoria, South Africa, and sequenced the msp1aS gene. The sequence of the OBP vaccine strain Msp1aS tandem repeat from 2014 was identical to that of the Israel strain (5) with four tandem repeats: Ac1 Ac1 Ac1 Ac2.
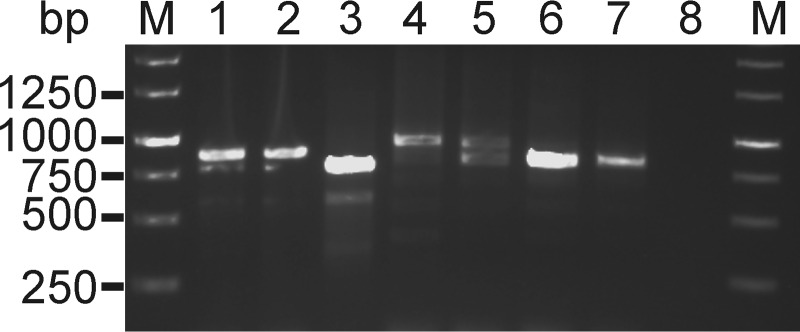
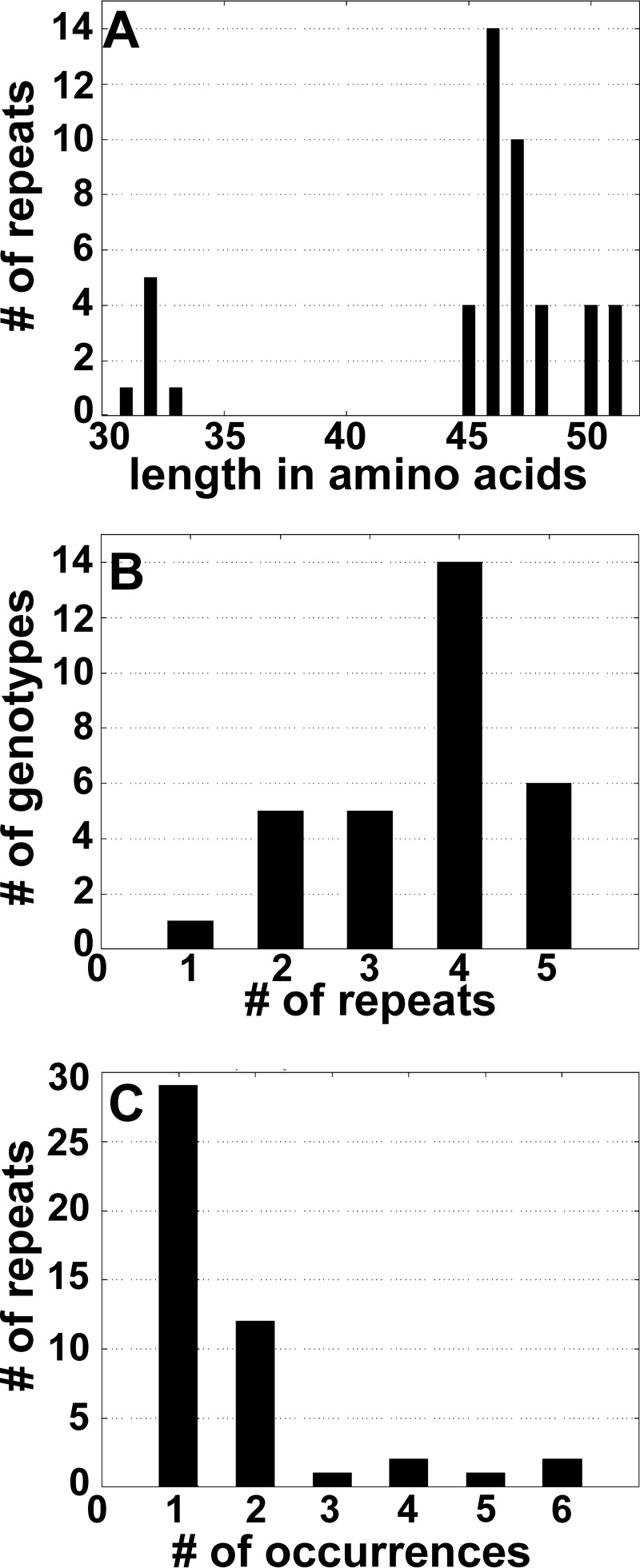
Based on the duplex qPCR results, A. marginale subsp. centrale-positive samples (n = 25) were selected for further analysis. Msp1aS primers amplified at least one single strong product from all samples tested. Some samples exhibited multiple bands which demonstrated mixed infection (Fig. 3). The msp1aS PCR products were cloned and sequenced, and sequence analyses confirmed the presence of tandem repeats similar to those of the vaccine strain (Table 2). The first five columns of Table 2 would combine to provide the full strain and sample designation as suggested previously (29), i.e., Ac11 Ac8_ZA, EC_2007_CNP_986; however, we have used shorter names for some of the genotypes for ease of discussion. The strains tested in this study yielded one to five repeat units as predicted from the PCR product sizes; however, there were strains that did not correspond with their PCR products (data not shown). Altogether, 47 different Msp1aS tandem repeats were identified. The repeats ranged from 45 to 51 amino acids with seven truncated repeats ranging from 31 to 33 amino acids (Fig. 4). The most common repeat length was 46 amino acids (Fig. 5A). The Ac1 and Ac2 tandem repeats, contained in the vaccine strain, were detected in cattle, buffalo, and wildebeest.
FIG 3.
Gel image showing amplicons of msp1aS. Lanes 1 and 2, vaccine strain (814 bp); lane 3, animal FS 383 (790 and 637 bp); lane 4, animal Berg10 (922 and 814 bp); lane 5, animal Berg12 (937 and 814 bp); lane 6, animal Berg20 (814 bp); lane 7, animal WC_108 (799 bp); lane 8, negative control. Note that for some samples only a subset of the amplicons were successfully sequenced, while for others, clones with different sequences were obtained from what appeared as a single band. Lanes marked “M” have a 1-kb molecular weight marker.
TABLE 2.
A. marginale subsp. centrale genotypes detected from South African bovine hosts (cattle, buffalo, and black wildebeest)
| Genotype | Country codea | Province codea | Yr | Animal no. | Sample clone ID | Host species | Origin, park, farm | Vaccine status | Size (bp) | No. of repeats | Short name |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ac1 Ac1 Ac1 Ac2 | IL | M | 2010 | Genome sequence | CP001759 | Cattle | Israel 2010 | + | 814 | 4 | Vaccine |
| Ac1 Ac1 Ac1 Ac2 | ZA | GP | 2014 | OBP vaccine | Cattle | OBP 2014b | + | 814 | 4 | Vaccine | |
| SANParks Biobanked samplesc | |||||||||||
| Ac11 Ac8 | ZA | EC | 2007 | CNP_986 | G | Buffalo | Cambedoo | − | 525 | 2 | |
| Ac9 Ac8 | ZA | EC | 2007 | CNP_986 | C | Buffalo | Cambedoo | − | 526 | 2 | |
| Ac11 Ac11 Ac11 Ac11 Ac8 | ZA | EC | 2007 | CNP_986 | C2 | Buffalo | Cambedoo | − | 940 | 5 | |
| Ac3 Ac4 Ac5 Ac6 | ZA | EC | 2007 | CNP_987 | J2 | Buffalo | Cambedoo | − | 823 | 3 | |
| Ac7 Ac8 | ZA | EC | 2007 | CNP_979 | D | Buffalo | Cambedoo | − | 526 | 2 | |
| Ac6 Ac35 Ac36T Ac37T Ac6 | ZA | NC | 2013 | MNP_999 | L | Buffalo | Mokala | − | 889 | 5 | |
| Ac38 Ac39T Ac34 Ac40T | ZA | NC | 2013 | MNP_999 | N | Buffalo | Mokala | − | 759 | 4 | |
| Ac38 Ac41T Ac42 Ac40T | ZA | NC | 2013 | MNP_1000 | A | Buffalo | Mokala | − | 733 | 4 | |
| Ac6Ac6 | ZA | NC | 2013 | MNP_1000 | G | Buffalo | Mokala | − | 790 | 2 | |
| Ac1 Ac1 Ac1 Ac2 | ZA | EC | 2013 | AEP_1003 | D | Buffalo | Addo | − | 814 | 4 | Vaccine |
| Ac7 Ac8 | ZA | EC | 2013 | AEP_1006 | D | Buffalo | Addo | − | 525 | 2 | |
| Ac38 Ac44T Ac43 | ZA | EC | 2013 | AEP_1006 | N | Buffalo | Addo | − | 628 | 3 | |
| Ac31 Ac8 | ZA | EC | 2013 | AEP_1006 | S | Buffalo | Addo | − | 526 | 2 | |
| Ac1 Ac1 Ac1 Ac1 | ZA | MP | 2008 | KNP_586 | A | Buffalo | Kruger | − | 814 | 4 | VV1 |
| Ac26 Ac26 Ac26 Ac2 | ZA | NC | 2011 | MNP_958 | F_w | Black wildebeest | Mokala | − | 862 | 4 | |
| Hluhluwe iMfolozi Park | |||||||||||
| Ac1 Ac1 Ac1 Ac2 | ZA | NL | 2008 | HiP_6 | 1 | Buffalo | Hluhluwe | − | 815 | 4 | Vaccine |
| Ac30 Ac24 Ac25 | ZA | NL | 2008 | HiP_6 | A | Buffalo | Hluhluwe | − | 940 | 3 | |
| Ac29 Ac29 Ac29 | ZA | NL | 2008 | HiP_6 | B | Buffalo | Hluhluwe | − | 703 | 3 | |
| Ac33 Ac3 Ac6 | ZA | NL | 2008 | HiP_6 | L | Buffalo | Hluhluwe | − | 691 | 3 | |
| NZG Biobanked samplesc | |||||||||||
| Ac20 Ac32 Ac21 Ac10 | ZA | WC | 2011 | WC_107 | E | Cattle | WC | − | 700 | 4 | |
| Ac1 Ac1 Ac1 Ac2 | ZA | WC | 2011 | WC _108 | A | Cattle | WC | − | 799 | 4 | Vaccine |
| Ac12 Ac12 Ac13 Ac13 Ac14 | ZA | NL | 2011 | KZN_138 | B | Cattle | NL | − | 919 | 5 | |
| Ac12 Ac12 Ac13 Ac13Ac14 | ZA | NL | 2011 | KZN_132 | A | Cattle | NL | − | 941 | 4 | |
| Ac12 Ac12 Ac13 Ac13 Ac14 | ZA | NL | 2011 | KZN_130 | B | Cattle | NL | − | 980 | 5 | |
| Ac15 Ac16 Ac16 Ac16 | ZA | FS | 2011 | FS_56 | B | Cattle | FS | − | 821 | 4 | |
| Ac16 Ac16 Ac16 | ZA | FS | 2011 | FS_383 | B | Cattle | FS | − | 637 | 3 | |
| Farm 1 | Cattle | ||||||||||
| Ac33 Ac3 Ac6 | ZA | NL | 2015 | Berg 10 | A | Cattle | Bergville | + | 691 | 3 | |
| Ac19 Ac19 Ac3 Ac6 | ZA | NL | 2015 | Berg 10 | G | Cattle | Bergville | + | 814 | 4 | |
| Ac17 Ac18 Ac45 Ac46T Ac47 | ZA | NL | 2015 | Berg 10 | J | Cattle | Bergville | + | 922 | 5 | |
| Ac1 Ac1 Ac1 Ac22 | ZA | NL | 2015 | Berg 12 | B | Cattle | Bergville | + | 811 | 4 | VV3 |
| Ac20 Ac21 Ac21 Ac20 | ZA | NL | 2015 | Berg 12 | E | Cattle | Bergville | + | 937 | 5 | |
| Ac1 Ac1 Ac1 Ac2 | ZA | NL | 2015 | Berg 12 | N | Cattle | Bergville | + | 814 | 4 | Vaccine |
| Ac23 Ac24 Ac25 Ac34 | ZA | NL | 2015 | Berg 19 | A | Cattle | Bergville | + | 940 | 5 | |
| Ac26 Ac12 Ac12 Ac27 Ac14 | ZA | NL | 2015 | Berg 19 | A_2 | Cattle | Bergville | + | 946 | 5 | |
| Ac1 Ac1 Ac1 Ac22 | ZA | NL | 2015 | Berg 19 | B | Cattle | Bergville | + | 811 | 4 | VV3 |
| Ac19 Ac3 Ac6 Ac6 | ZA | NL | 2015 | Berg 19 | I | Cattle | Bergville | + | 826 | 4 | |
| Farm 2 | |||||||||||
| Ac1 Ac1 Ac1 Ac22 | ZA | NL | 2015 | Berg 25 | A | Cattle | Bergville | + | 814 | 4 | VV3 |
| Ac1 | ZA | NL | 2015 | Berg 25 | E | Cattle | Bergville | + | 391 | 1 | |
| Ac1 Ac1 Ac1 Ac2 | ZA | NL | 2015 | Berg 25 | B | Cattle | Bergville | + | 814 | 4 | Vaccine |
| Ac1 Ac1 Ac1 Ac1 | ZA | NL | 2015 | Berg 25 | E_2 | Cattle | Bergville | + | 814 | 4 | VV1 |
| Ac1 Ac1 Ac1 Ac2 Ac2 | ZA | NL | 2015 | Berg 25 | X | Cattle | Bergville | + | 914 | 5 | VV2 |
| Ac1 Ac1 Ac1 Ac2 | ZA | NL | 2015 | Berg 27 | D | Cattle | Bergville | + | 814 | 4 | Vaccine |
| Ac1 Ac1 Ac1 Ac1 | ZA | NL | 2015 | Berg 27 | E | Cattle | Bergville | + | 956 | 5 | VV1 |
| Ac1 Ac1 Ac1 Ac2 Ac2 | ZA | NL | 2015 | Berg 27 | B | Cattle | Bergville | + | 955 | 5 | VV2 |
| Ac1 Ac1 Ac1 Ac2 | ZA | NL | 2015 | Berg 17 | A | Cattle | Bergville | + | 943 | 5 | Vaccine |
| Ac1 Ac1 Ac1 Ac2 | ZA | NL | 2015 | Berg 24 | A | Cattle | Bergville | + | 814 | 5 | Vaccine |
| Ac1 Ac1 Ac1 Ac2 Ac2 | ZA | NL | 2015 | Berg 24 | C | Cattle | Bergville | + | 955 | 5 | VV2 |
| Ac1 Ac28 Ac2 Ac28 | ZA | NL | 2015 | Berg 24 | V | Cattle | Bergville | + | 814 | 4 | |
| Ac1 Ac1 Ac1 Ac2 | ZA | NL | 2015 | Berg 30 | G | Cattle | Bergville | + | 811 | 4 | Vaccine |
| Ac1 Ac1 Ac1 Ac2 Ac2 | ZA | NL | 2015 | Berg 30 | I | Cattle | Bergville | + | 954 | 5 | VV2 |
| Ac1 Ac1 Ac1 Ac1 | ZA | NL | 2015 | Berg 20 | H3 | Cattle | Bergville | + | 814 | 4 | VV1 |
Country and province abbreviations follow ISO 3166-2.
OBP, Onderstepoort Biological Products (Pretoria, South Africa), which produces A. marginale subsp. centrale vaccine for sale.
SANParks, South African National Parks; NZG, National Zoological Gardens of South Africa Biobanks.
FIG 4.
Alignment of A. marginale subsp. centrale Msp1aS tandem repeats detected from South African cattle, buffalo, and black wildebeest. The 47 repeat types were aligned using the AlignX module of Vector NTI, and groups of identical amino acids are highlighted on a black background. Ac1 and Ac2, the repeats present in the vaccine strain, are indicated with an asterisk.
FIG 5.
Metrics for A. marginale subsp. centrale Msp1aS repeats. (A) Number of repeats with a given number of amino acids; i.e., there are four repeats with a length of 45 amino acids. (B) Number of genotypes having a given number of repeats; i.e., 14 genotypes contain four repeats. (C) Number of times a given repeat occurs in our genotype data set; i.e., two repeats occur in six different genotypes.
The vaccine strain was detected in cattle from Bergville which were previously vaccinated with A. marginale subsp. centrale vaccine. We tested six cattle from Bergville farm 2 which yielded 15 msp1aS sequences. The vaccine genotype was detected in five of the six cattle (Table 2). Interestingly, two “vaccine variant” genotypes were detected that were closely related to the vaccine strain genotype and differed by only a single amino acid (VV1 and VV3). Another vaccine variant genotype, VV2 (Ac1 Ac1 Ac1 Ac2 Ac2), that had one additional Ac2 repeat but was otherwise identical to the vaccine strain genotype was noted. Two additional genotypes that were less obviously related to the vaccine strain were detected. Three cattle were tested on Bergville farm 1, resulting in 10 msp1aS sequences. Interestingly, the vaccine genotype was only detected in one of these animals despite the fact that these animals were reported as being vaccinated, while two animals contained the related genotype VV3. Seven additional genotypes were detected on farm 1 that were not closely related to the vaccine genotype.
Interestingly, the vaccine genotype and one of the vaccine variant genotypes were also detected in unvaccinated animals, including buffalo (HIP_6, AEP_1003, and KNP_586) and cattle (WC_108). Genotype Ac33 Ac3 Ac6 was detected in a buffalo from Hluhluwe National Park and in a cow from Bergville farm 1. Several truncated repeats were detected (i.e., Ac36T), and although these predominated in the buffalo samples, a genotype containing a truncated repeat was also detected on Bergville farm 1.
RepeatAnalyzer is a program we developed recently to house, curate, and provide metrics for repeat sequences used to characterize bacteria (29). In the present study, we applied it to the analysis of msp1aS repeats. The most common genotype structure we detected contained four repeats, with genotypes having from one to five repeats (Fig. 5B). Most repeats occurred only once with two repeats being detected in six different genotypes (Ac1 and Ac6) (Fig. 5C). The Ac1 repeat is not only detected in the vaccine strain but also in several vaccine variant genotypes that were detected on Bergville farm 2. The Ac6 repeat was prevalent in genotypes detected in wildlife and, interestingly, was also detected in genotypes found on Bergville farm 1. In general, we found that the average number of amino acid changes (edit distance) between any two A. marginale subsp. centrale repeats was high (13.7) and was normally distributed, with 97.8% of data falling within 2 standard deviations. There was a mean of 0.9 and 1.4 repeats at an edit distance of 1 and 2, respectively, from any given repeat. Despite the high level of variation between repeats, we found five repeats within an edit distance of two from Ac1 (Ac2, Ac26, Ac12, Ac20, and Ac48) and seven repeats within two edits of Ac2 (Ac1, Ac14, Ac28, Ac15, Ac16, Ac22, and Ac26).
Diversity analysis and repeat distribution.
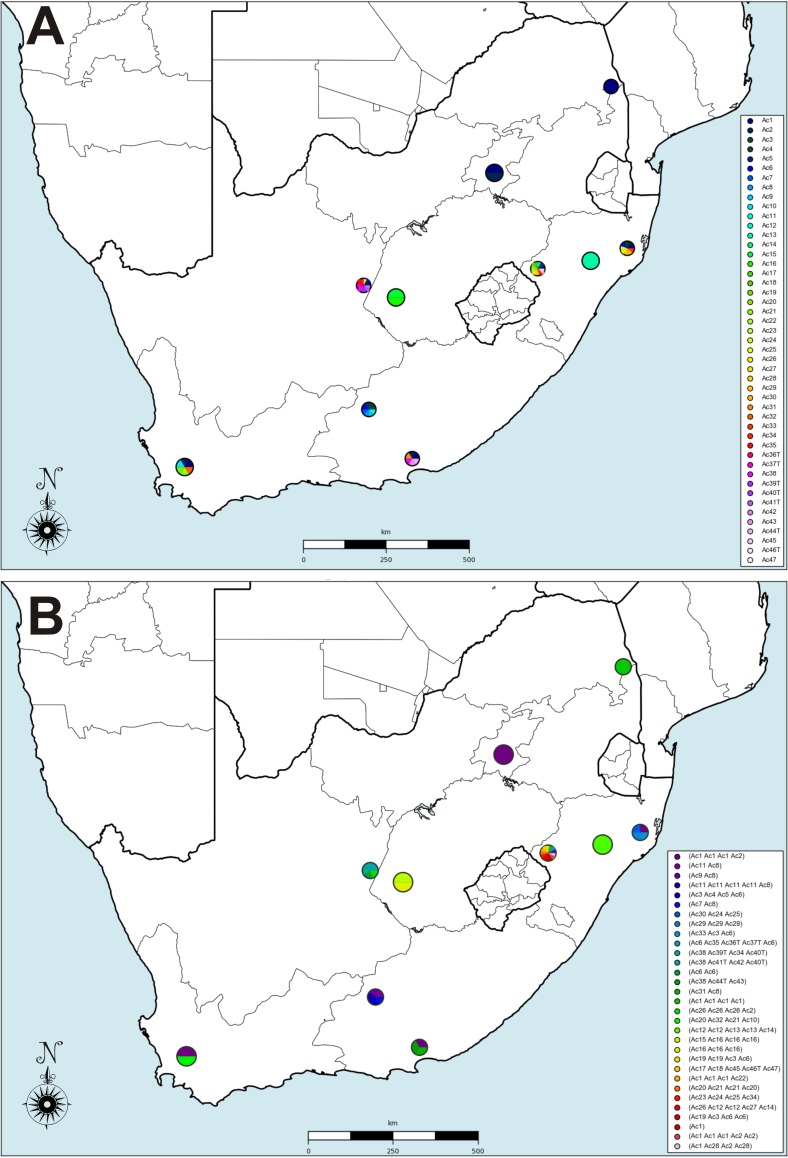
Using RepeatAnalyzer, we see that South Africa has a large number of unique A. marginale subsp. centrale repeats (Table 3, GDM1-L), while having an intermediate amount of repeat diversity in general (Table 3, GDM1-G). There is a higher diversity of repeats among the samples isolated from buffalo hosts than among those from cattle hosts, although this would be expected as many of the cattle were vaccinated and would be expected to exhibit the same repeat structure as the vaccine strain. GDM2 measures how uniformly the repeat occurrences in the strains in a region (local) or the region as a whole (global) are distributed. For both GDM2 metrics, the South African values are low, indicating that the repeats are dispersed; i.e., there is not a preponderance of a single repeat type in individual strains or for the country as a whole. The GDM2 values are higher for cattle than for buffalo-derived samples, reflecting more uniformity in the repeats detected in samples from cattle than from buffalo. When examining whether repeats and strains occur in multiple provinces, we have msp1aS data from seven of South Africa's nine provinces (Fig. 6). The repeats and strains are mapped according to GPS coordinates, so multiple locations within a province can be visualized and distinguished. Several repeats were detected in multiple locations (Fig. 6A). Repeats Ac1 and Ac2 were found in Mpumalanga, Gauteng, KwaZulu-Natal, Eastern Cape, and Western Cape provinces. The vaccine strain is detected in cattle from KwaZulu-Natal, the Eastern Cape, and the Western Cape (Fig. 6B), which is interesting as we tested vaccinated animals only in KwaZulu-Natal. Gauteng also shows positive for the vaccine strain, but this is due to the purchased vaccine itself.
TABLE 3.
Diversity scores for cattle and wildlife hosts by province and host
| Location | GDM1-L | GDM1-G | GDM2-L | GDM2-G |
|---|---|---|---|---|
| All | 0.747 | 0.420 | 0.065 | 0.022 |
| Eastern Cape | 0.863 | 0.583 | 0.069 | 0.060 |
| Gauteng | 0.500 | 0.500 | 0.250 | 0.250 |
| KwaZulu-Natal | 0.696 | 0.419 | 0.071 | 0.044 |
| Mpumalanga | 0.250 | 0.250 | 0 | 0 |
| Northern Cape | 0.760 | 0.632 | 0.067 | 0.050 |
| Free State | 0.417 | 0.286 | 0.125 | 0.357 |
| Western Cape | 0.750 | 0.750 | 0.125 | 0.093 |
| Buffalo | 0.781 | 0.500 | 0.051 | 0.030 |
| Cattle | 0.684 | 0.418 | 0.081 | 0.041 |
FIG 6.
Maps of repeat and strain distribution. (A) Repeats mapped to the provinces of South Africa by GPS coordinates. (B) Strain genotypes mapped to the provinces of South Africa by GPS coordinates. The size of the circle indicates the precision of the location report, with three sizes being possible, corresponding to country, province, and precise GPS location. In these maps, there are no reports that are simply to the country level; i.e., all locations are at the provincial level or more specific. Therefore, there are only two sizes of circles shown. The samples collected from the Free State and Western Cape are marked at the provincial level and, thus, have larger markers.
DISCUSSION
We tested animals from several different parks and farms and showed that A. marginale subsp. centrale infection is prevalent in black and blue wildebeest, eland, buffalo, waterbuck, and cattle. A. marginale subsp. centrale has rarely been examined on its own, as typically researchers/ranchers are interested in A. marginale infection, and the competitive enzyme-linked immunosorbent assay (cELISA) often used for detection does not discriminate between A. marginale and A. marginale subsp. centrale infection. One study using the cELISA showed high seroprevalence of Anaplasma spp. in wildlife from Kenya with eland and blue wildebeest testing at 100% and 96%, respectively. With a reverse line blot assay, it was shown that Anaplasma spp. are prevalent in buffalo in northern Botswana with A. marginale subsp. centrale being the most prevalent (22). This suggests that wildlife species are reservoirs of A. marginale subsp. centrale.
We examined positive samples for msp1aS genotype, a genotyping scheme that has not previously been employed for A. marginale subsp. centrale. We identified 47 Msp1aS repeats which corresponded to 32 A. marginale subsp. centrale genotypes detected in cattle, buffalo, and wildebeest. The most common A. marginale subsp. centrale genotype among cattle samples was the vaccine genotype. This is not surprising as both farms that we sampled previously vaccinated with A. marginale subsp. centrale vaccine purchased from OBP. It is worth noting that cattle from farm 1 graze together with goats, sheep, and reedbuck, which might explain the diversity of A. marginale subsp. centrale strains detected on farm 1. We speculate that there is circulation of A. marginale subsp. centrale strains among different hosts, which led to the variety of genotypes detected on this farm. Cattle from farm 2 are confined within a grazing area with no interaction with other ruminants. The vaccine genotype was detected in all but one of the animals tested on this farm. In addition to the vaccine genotype, several closely related genotypes were detected, which suggests that the vaccine genotype is changing under selection pressure. This is interesting as we do not see these types of changes in the msp1α genotype in A. marginale-infected cattle. All repeats detected on farm 2 had an edit distance of two or less from one of the vaccine strain repeats, indicating that these repeats were closely related to the vaccine strain repeats. However, we cannot be sure that the vaccine strain is changing rather than there being an introduction of these new, related genotypes.
The unvaccinated cattle samples from Western Cape and Free State had different A. marginale subsp. centrale genotypes, while unvaccinated cattle samples from KwaZulu-Natal all had the same A. marginale subsp. centrale genotype. The vaccine strain was detected in one of the unvaccinated cattle in the Western Cape. The A. marginale subsp. centrale genotypes obtained from wild ruminants were diverse, demonstrating geographic segregation of national parks. The repeat Ac8 was common in the msp1aS genotypes found in buffalo, even though the buffalo were sourced from parks distributed around South Africa. Ac8 has an edit distance of nine to both repeats Ac1 and Ac2, indicating that it is not closely related to the vaccine strain repeats.
While we have presented diversity metrics broken down by province, we think that the sample size is too small for this to be really meaningful in most cases, i.e., in Mpumalanga and Gauteng, there is an n = 1. More importantly, these metrics show us that for South Africa, as a whole, there is a high degree of repeat diversity within genotypes (Table 3, GDM1-L) and a moderate degree of novel genotypes across the country (Table 3, GDM1-G). The low GDM2 values indicate that the repeats are dispersed, which is what is expected when the numbers of unique repeats and genotypes are high. This high degree of novel repeats indicates that the repeats have likely been circulating in nature and undergoing selection and change separate from the vaccine strain. As more data are collected, it will be interesting to see if these metrics shift and how these metrics compare with those collected in other countries.
While the A. marginale subsp. centrale vaccine strain was thought for a long time not to be transmitted by most ticks, it was shown that, in fact, it colonized the tick well but was not secreted into the tick saliva in sufficient quantities for robust transmission (23, 24). Dramatically increasing tick numbers in transmission experiments overcame the transmission barrier (25). Is the reduced ability of the A. marginale subsp. centrale vaccine strain to be tick transmitted due to long serial needle passage through cattle? Or is there, perhaps, a specific vector-pathogen adaptation? There is a report of apparently efficient tick transmission of A. marginale subsp. centrale vaccine strain from Rhipicephalus simus ticks (26). Although R. simus is a proven vector in laboratory conditions, this tick is not found on cattle in large numbers, and the immature stages do not normally infest cattle (27). It would appear that the strains that we have detected circulating in wild animals today are maintained in nature via a natural tick-transmission cycle; however, this remains a speculation at this stage, as we have not tested ticks or performed transmission studies due to the complexities of working with the ecosystem of infections present in South Africa. If, in fact, A. marginale subsp. centrale is being spread through natural transmission to cattle, it is likely mitigating some of the disease burden of anaplasmosis caused by A. marginale.
In conclusion, this paper presents a novel genetic test based on msp1aS to discriminate strains of A. marginale subsp. centrale and shows that the vaccine strain is found widely distributed across South Africa and in animals that do not have a history of vaccination. Further, we present metrics indicating a high degree of Msp1aS repeat diversity in South Africa. Our results indicate the significance of wildlife as reservoir hosts for A. marginale subsp. centrale.
ACKNOWLEDGMENTS
We thank B. Christoff from Plaas Hongerspoort and A. Shepherd from Tugela Veterinary Clinic, Bergville, KwaZulu-Natal, who helped us collect blood samples from vaccinated cattle and M.S. Mtshali and A. Mutshembele for providing blood samples from unvaccinated cattle from KwaZulu-Natal, Western Cape, and Free State. Wildlife samples were obtained from the South African National Parks SANParks Biobank under reference number LARBJ1118 Conservation Genetics and from the Wildlife Biological Resource Center (WBRC) and Biobank SA under the auspices of the National Zoological Gardens (NZG) of South Africa and Johannesburg Zoo. We thank Dave Cooper for providing buffalo blood samples from Hluhluwe-iMfolozi Park. We thank Erich Zweygarth for Anaplasma sp. Omatjenne.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Any opinion, finding, conclusion, or recommendation expressed in this material is that of the authors, and the NRF does not accept any liability in this regard.
REFERENCES
- 1.Theiler A. 1910. Anaplasma marginale: the marginal points in the blood of cattle suffering from a specific disease. Report of the government veterinary bacteriologist, 1908-1909. Transvaal, South Africa. [Google Scholar]
- 2.Losos GJ. 1986. Anaplasmosis, p 743−795. In Infectious tropical diseases of domestic animals. Longman House, Essex, United Kingdom. [Google Scholar]
- 3.Theiler A. 1911. Further investigations into anaplasmosis of South African cattle, p 7−46. First Report of the Director of Veterinary Research, Union of South Africa. Johannesburg, South Africa. [Google Scholar]
- 4.Aubry P, Geale DW. 2011. A review of bovine anaplasmosis. Transbound Emerg Dis 58:1–30. doi: 10.1111/j.1865-1682.2010.01173.x. [DOI] [PubMed] [Google Scholar]
- 5.Herndon DR, Palmer GH, Shkap V, Knowles DP Jr, Brayton KA. 2010. Complete genome sequence of Anaplasma marginale subsp. centrale. J Bacteriol 192:379–380. doi: 10.1128/JB.01330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuttler KL. 1984. Anaplasma infections in wild and domestic ruminants: a review. J Wildl Dis 20:12–20. doi: 10.7589/0090-3558-20.1.12. [DOI] [PubMed] [Google Scholar]
- 7.Anziani OS, Tarabla HD, Ford CA, Galleto C. 1987. Vaccination with Anaplasma centrale: Response after an experimental challenge with Anaplasma marginale. Trop Anim Health Prod 19:83–87. doi: 10.1007/BF02297324. [DOI] [PubMed] [Google Scholar]
- 8.Neitz WO. 1935. Bovine anaplasmosis: the transmission of Anaplasma marginale to a black wildebeest (Connochaetes gnou). Onderstepoort J Vet Sci Anim Ind 5:9–11. [Google Scholar]
- 9.Potgieter FT. 1979. Epizootiology and control of anaplasmosis in South Africa. J S Afr Vet Assoc 50:367–372. [PubMed] [Google Scholar]
- 10.Smith RD, Woolf A, Hungerford LL, Sundberg JP. 1982. Serologic evidence of Anaplasma marginale infection in Illinois white-tailed deer. J Am Vet Med Assoc 181:1254–1256. [PubMed] [Google Scholar]
- 11.Potgieter FT, Stoltsz W. 2004. Bovine anaplasmosis, p 598–600. In Coetzer JAW, Tustin RC (ed), Infectious diseases of livestock, 2nd ed, vol 1 Oxford University Press, Cape Town, South Africa. [Google Scholar]
- 12.Georges K, Loria GR, Riili S, Greco A, Caracappa S, Jongejan F, Sparagano O. 2001. Detection of haemoparasites in cattle by reverse line blot hybridisation with a note on the distribution of ticks in Sicily. Vet Parasitol 99:273–286. doi: 10.1016/S0304-4017(01)00488-5. [DOI] [PubMed] [Google Scholar]
- 13.Carelli G, Decaro N, Lorusso E, Paradies P, Elia G, Martella V, Buonavoglia C, Ceci L. 2008. First report of bovine anaplasmosis caused by Anaplasma centrale in Europe. Ann N Y Acad Sci 1149:107–110. doi: 10.1196/annals.1428.069. [DOI] [PubMed] [Google Scholar]
- 14.Allred DR, McGuire TC, Palmer GH, Leib SR, Harkins TM, McElwain TF, Barbet AF. 1990. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc Natl Acad Sci U S A 87:3220–3224. doi: 10.1073/pnas.87.8.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowie MV, de la Fuente J, Kocan KM, Blouin EF, Barbet AF. 2002. Conservation of major surface protein 1 genes of Anaplasma marginale during cyclic transmission between ticks and cattle. Gene 282:95–102. doi: 10.1016/S0378-1119(01)00845-9. [DOI] [PubMed] [Google Scholar]
- 16.de la Fuente J, Ruybal P, Mtshali MS, Naranjo V, Shuqing L, Mangold AJ, Rodríguez SD, Jiménez R, Vicente J, Moretta R, Torina A, Almazán C, Mbati PM, de Echaide ST, Farber M, Rosario-Cruz R, Gortazar C, Kocan KM. 2007. Analysis of world strains of Anaplasma marginale using major surface protein 1a repeat sequences. Vet Microbiol 119:382–390. doi: 10.1016/j.vetmic.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Mutshembele AM, Cabezas-Cruz A, Mtshali MS, Thekisoe OMM, Galindo RC, de la Fuente J. 2014. Epidemiology and evolution of the genetic variability of Anaplasma marginale in South Africa. Ticks Tick Borne Dis 5:624–631. doi: 10.1016/j.ttbdis.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Mtshali MS, de la Fuente J, Ruybal P, Kocan KM, Vicente J, Mbati PA, Shkap V, Blouin EF, Mohale NE, Moloi TP, Spickett AM, Latif AA. 2007. Prevalence and genetic diversity of Anaplasma marginale strains in cattle in South Africa. Zoonoses Public Health 54:23–30. doi: 10.1111/j.1863-2378.2007.00998.x. [DOI] [PubMed] [Google Scholar]
- 19.Decaro N, Carelli G, Lorusso E, Lucente MS, Greco G, Lorusso A, Radogna A, Ceci L, Buonavoglia C. 2008. Duplex real-time polymerase chain reaction for simultaneous detection and quantification of Anaplasma marginale and Anaplasma centrale. J Vet Diagn Invest 20:606–611. doi: 10.1177/104063870802000511. [DOI] [PubMed] [Google Scholar]
- 20.Carelli G, Decaro N, Lorusso A, Elia G, Lorusso E, Mari V, Ceci L, Buonavoglia C. 2007. Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet Microbiol 124:107–114. doi: 10.1016/j.vetmic.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 21.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eygelaar D, Jori F, Mokopasetso M, Sibeko KP, Collins NE, Vorster I, Troskie M, Oosthuizen MC. 2015. Tick-borne haemoparasites in African buffalo (Syncerus caffer) from two wildlife areas in Northern Botswana. Parasit Vectors 8:26. doi: 10.1186/s13071-014-0627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueti MW, Reagan JO Jr, Knowles DP Jr, Scoles GA, Shkap V, Palmer GH. 2007. Identification of midgut and salivary glands as specific and distinct barriers to efficient tick-borne transmission of Anaplasma marginale. Infect Immun 75:2959–2964. doi: 10.1128/IAI.00284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shkap V, Kocan K, Molad T, Mazuz M, Leibovich B, Krigel Y, Michoytchenko A, Blouin E, De la Fuente J, Samish M. 2009. Experimental transmission of field Anaplasma marginale and the A. centrale vaccine strain by Hyalomma excavatum, Rhipicephalus sanguineus and Rhipicephalus (Boophilus) annulatus ticks. Vet Microbiol 134:254–260. doi: 10.1016/j.vetmic.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Ueti MW, Knowles DP, Davitt CM, Scoles GA, Baszler TV, Palmer GH. 2009. Quantitative differences in salivary pathogen load during tick transmission underlie strain-specific variation in transmission efficiency of Anaplasma marginale. Infect Immun 77:70–75. doi: 10.1128/IAI.01164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potgieter FT, van Rensburg L. 1987. Tick transmission of Anaplasma centrale. Onderstepoort J Vet Res 54:5–7. [PubMed] [Google Scholar]
- 27.Potgieter FT. 1981. Tick transmission of anaplasmosis in South Africa, p 222 In Proceedings of the International Conference on Tick Biology and Control, Grahamstown, South Africa. [Google Scholar]
- 28.Chaisi ME, Baxter JR, Hove P, Choopa CN, Oosthuizen MC, Brayton KA, Khumalo ZTH, Mutshembele A, Mtshali S, Collins NE. Comparison of molecular tests for the detection of Anaplasma marginale and A. centrale in South Africa. Onderstepoort J Vet Res, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catanese HN, Brayton KA, Gebremedhin AH. 2016. RepeatAnalyzer: a tool for analysing and managing short-sequence repeat data. BMC Genomics 17:422. doi: 10.1186/s12864-016-2686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]