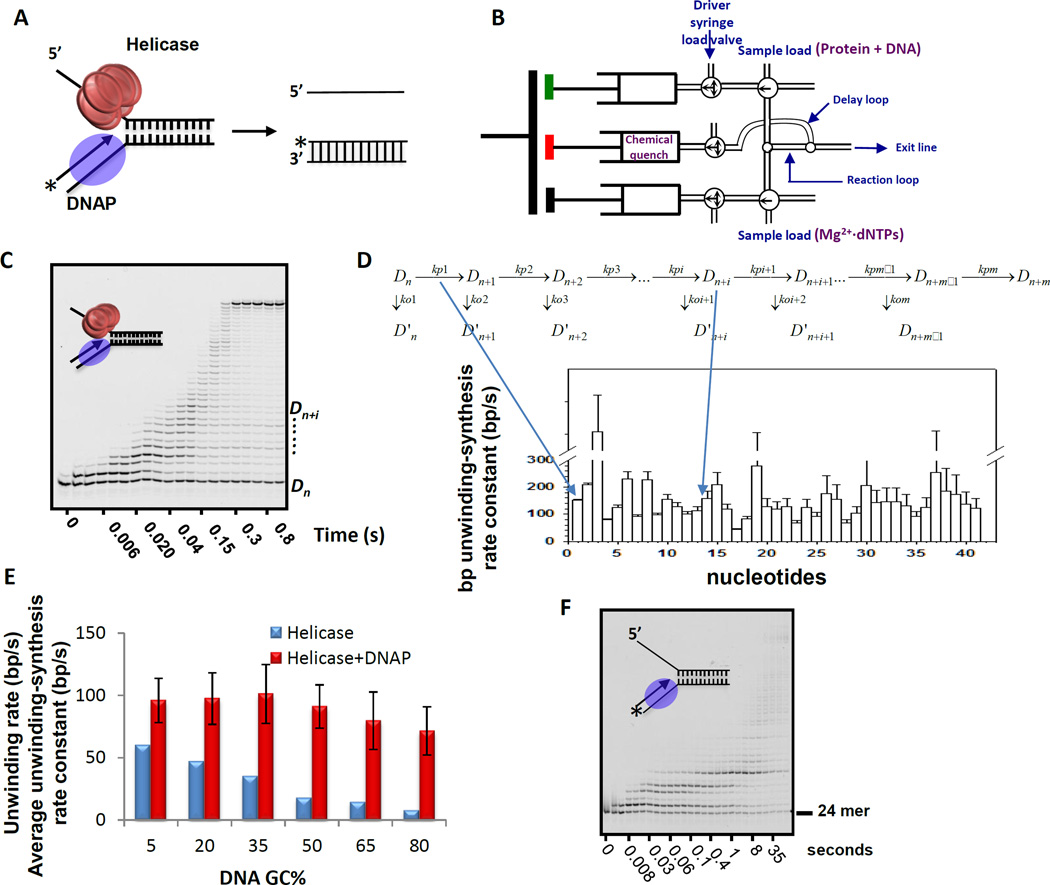
Figure 3. Gel-based assay to determine the base pair unwinding-synthesis rate constant of the helicase-DNAP complex.
(A) The helicase-DNAP complex is assembled on the replication fork DNA. The 5’-end of the 24-mer primer in the fork DNA is labeled with a fluorophore or γ[32Pi]. The unwinding-synthesis reaction initiated with dNTPs is monitored by following the extension of the labeled DNA primer. (B) Schematic of the rapid quench-flow instrument for rapid mixing and quenching of the reactions with millsecond time resolution. The duration of the reaction is changed by adjusting the volume of the delay line and the flow rate through the delay loop. (C) Image of polyacrylamide/urea sequencing gel showing extension of the 24-mer DNA primer by T7 helicase-DNAP (1 mM dNTPs) at 18 °C. The T7 helicase is the product of T7 gp4, which contains both helicase and primase activities[58]. T7 DNAP is a 1:1 complex of polymerase T7 gp5 and processivity factor E. coli thioredoxin[59]. (D) The polymerization model is used to globally fit the formation and decay of each primer extension product to obtain the individual base pair unwinding-synthesis rate constants. The kp1, kp2, etc. are the rate constants for the formation of Dn+1, Dn+2, etc. extension products, respectively. The ko1, ko2, etc. are the rate constants for the dissociation of the DNAP from the Dn+1, Dn+2, etc. complex intermediates. The computer fitting provides the rate constants for the individual nucleotide addition reactions (kp1, kp2, etc.) from which one can calculate the average single base pair unwinding synthesis rate constant. (E) The average unwinding-synthesis rate constant of the helicase-DNAP is compared to the helicase’s rate constant of unwinding on fork DNA substrates with 5–80% GC-content. (F) The sequencing gel image shows the strand-displacement DNA synthesis activity of the isolated T7 DNAP (1 mM dNTPs) at 18 °C.