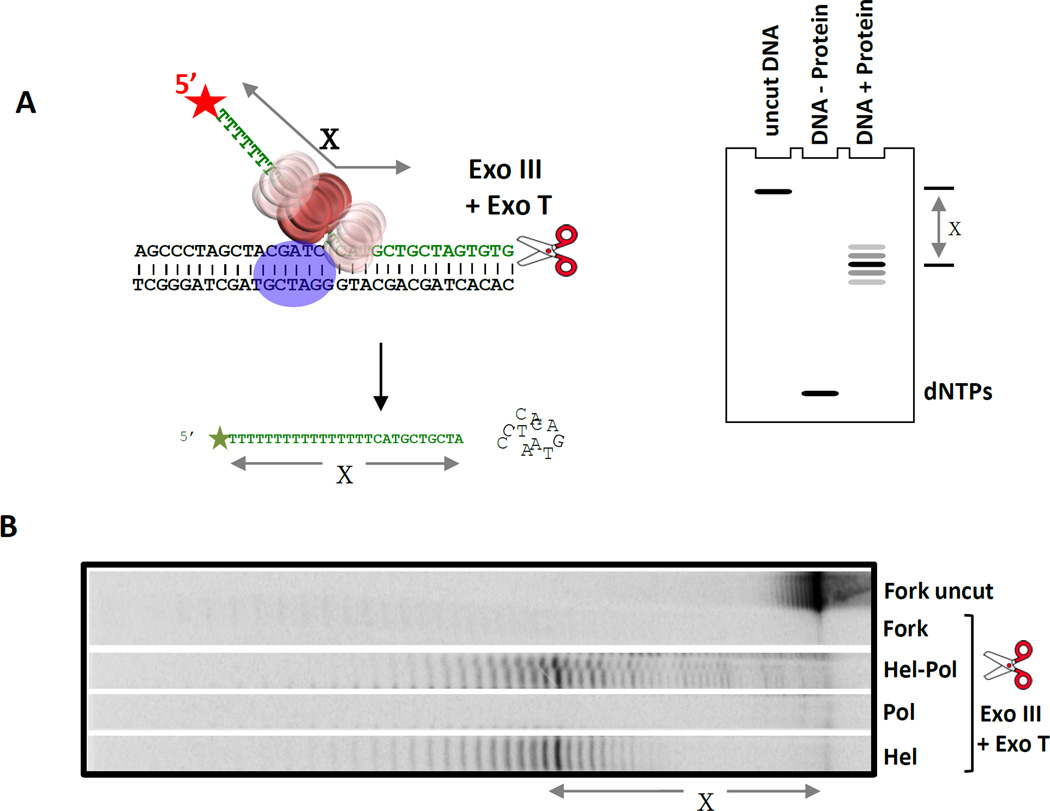
Figure 6. Exonuclease footprinting to determine the position of T7 helicase on the replication fork substrate in complex with T7 DNAP.
(A) The 5’ end of the lagging DNA strand in the fork DNA (with a 36-nt ssDNA tail) is labeled to visualize the protected DNA fragments after digestion with a combination of Exo III and Exo T enzymes. The simulated gel cartoon shows the expected length of the protected DNA fragments when the helicase is bound at the fork junction. The length of the protected DNA fragments (‘X’) is determined by analysis on a high resolution sequencing gel. If X is less than 36-nt, then the helicase is positioned away from the fork junction, if X is 36-nt then the downstream boundary of the helicase is at the fork junction, and if X is greater than 36-nt then the helicase is positioned downstream of the fork junction. (B) Sequencing gel shows the products of ExoIII + ExoT digestion (X = 38-nt) in the presence and absence of proteins (T7 helicase and T7 helicase-DNAP). The length of the protected DNA fragments with the helicase and helicase-DNAP are similar indicating that the position of the helicase on the fork DNA does not change in the presence of the DNAP. The prominent 38-nt central band indicates that the helicase is within 2-nt downstream of the fork junction [21].