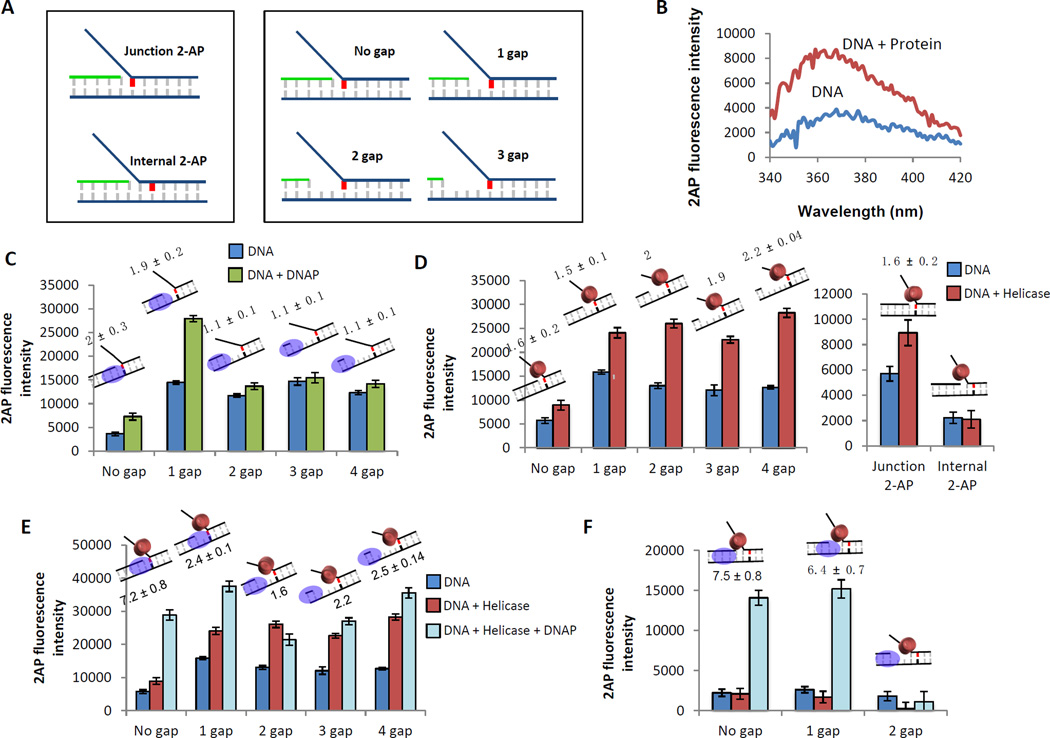
Figure 8. 2-AP fluorescence assay to determine the relative contribution of the helicase and DNAP to dsDNA unwinding.
(A) Schematics of the 2-AP labeled fork DNA substrates. A single 2-AP probe (indicated in red) is placed at the fork junction or one base pair downstream of the fork junction. The primer end is adjusted to generate fork DNA substrates with no gap, 1 gap, 2 gap, or 3 nt ssDNA gap between the primer end and fork junction. (B) Change in 2-AP fluorescence intensity on protein binding is monitored over a range of wavelengths. The change in 2-AP intensity on protein binding reflects on the effect of the protein on the 2-AP base pair. (C) Fluorescence intensities of 2-AP modified replication fork DNA with (green bars) and without (blue bars) T7 DNAP. Cartoons of the DNA substrate used in the experiment and the fold change of fluorescence intensity on protein binding are shown above each bar. (D) Fluorescence intensities of 2-AP modified replication fork DNA with (red bars) and without (blue bars) T7 helicase. (E) Fluorescence intensities of 2-AP modified replication fork DNA with (light blue bars) and without (blue bars) T7 helicase-DNAP. The red bars show intensity changes with T7 helicase alone. (F) Fluorescence intensities of 2-AP modified replication fork DNA with (light blue bars) and without (blue bars) T7 helicase-DNAP. The red bars are the effect of just T7 helicase binding in the absence of T7 DNAP.