Figure 1.
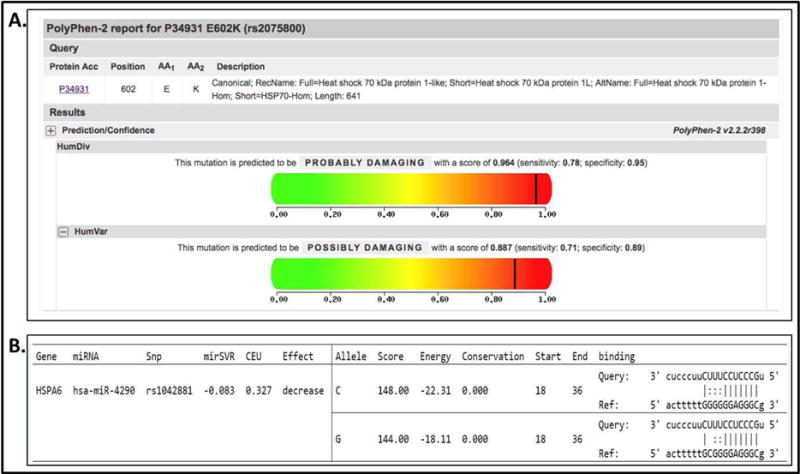
Results of HSPA1L and HSPA6 in silico analysis of SNP function. A, HSPA1L rs2075800 results in a glutamine to lysine substitution at position 602 of the protein (E602K) that is considered potentially damaging; B, HSPA6 rs1042881 located in the 3′ UTR is predicted to have different binding affinity to the microRNA hsa-miR-4290. Albeit not significant, the C allele appears to have increased affinity to hsa-miR-4290.
