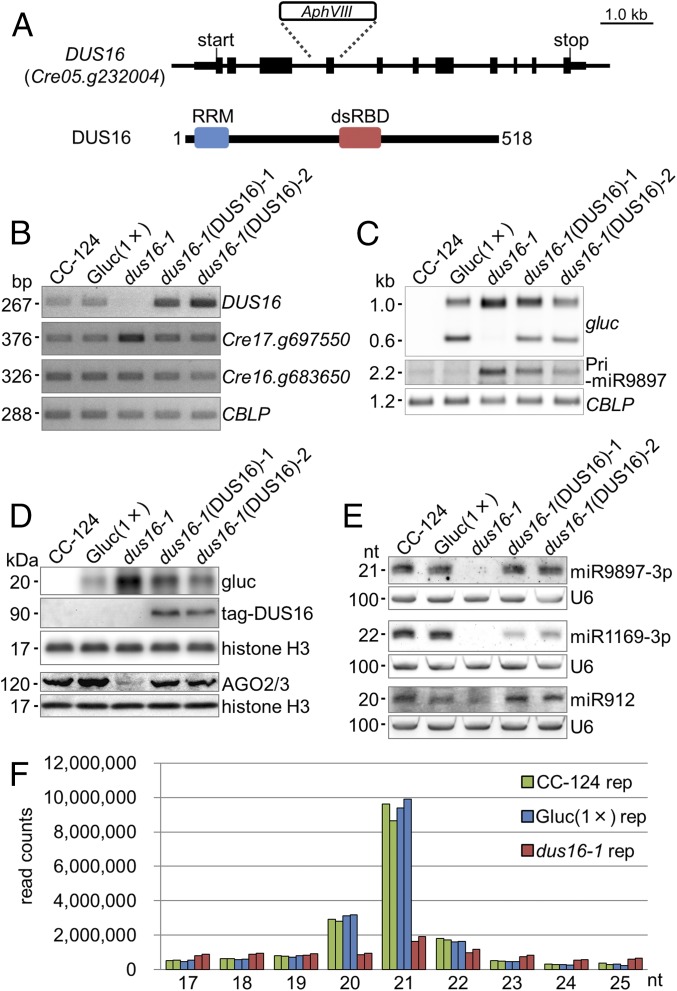
Fig. 1.
Characterization of the DUS16 mutant. CC-124, wild-type strain; Gluc(1×), CC-124 transgenic strain expressing Gaussia luciferase (gluc) bearing a sequence perfectly complementary to Chlamydomonas miR9897-3p; dus16-1, DUS16-defective mutant of Gluc(1×); dus16-1(DUS16)-1 and -2, strains expressing HA-FLAG-tagged DUS16 in a dus16-1 background. (A) Schematic diagram of the gene structure and transcript of DUS16 (Cre05.g232004). dsRBD, dsRNA-binding domain; RRM, ssRNA-binding domain. (B–E) Each panel is representative of three independent experiments. (B) RT-PCR analysis in the indicated strains. Cre17.g697550 is a target of miR B-mediated cleavage. Cre16.g683650 is a target of miR C-mediated translation repression. A housekeeping gene, CBLP, was used as a control. (C) Northern blotting of gluc mRNA and the pri-miR9897 transcript in the indicated strains. Detection of CBLP was performed to check for equivalent loading of the RNA samples. (D) Immunoblotting of gluc, tagged DUS16, and AGO2/3 proteins in the indicated strains. The tagged DUS16 protein was detected with a monoclonal antibody against the HA tag. Both AGO2 and AGO3 polypeptides are recognized by a polyclonal antibody. Detection of histone H3 was performed to confirm similar loading of the protein samples. (E) Northern blotting of Chlamydomonas mature miRNAs in the indicated strains. U6 small nuclear RNA was used as a control for equal loading of the RNA samples. (F) Small RNA-seq analysis of CC-124 (green), Gluc(1×) (blue), and dus16-1 (red). Biological duplicates of sRNA libraries were generated from the indicated strains. sRNA reads, 17–25 nt in length, were classified according to their size. Absolute read counts, without normalization, are indicated.