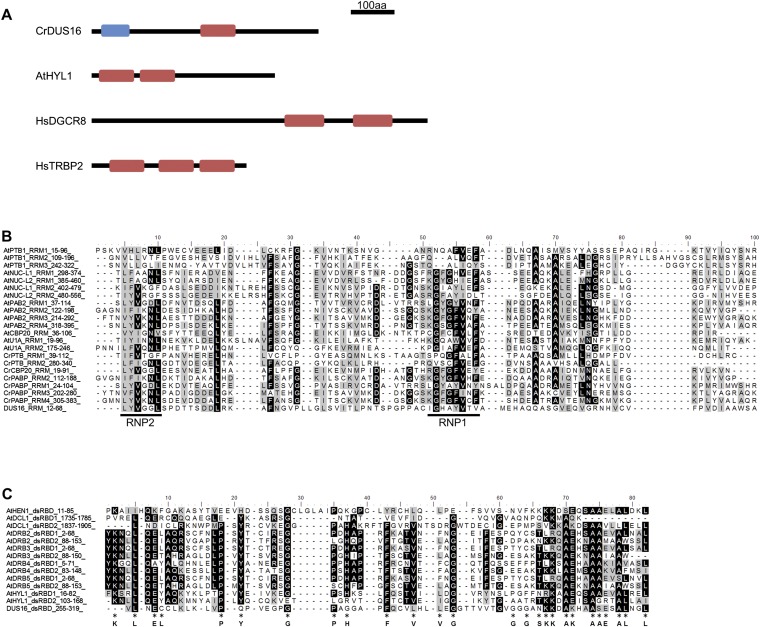
Fig. S1.
Domain structure of DUS16, HYL1, DGCR8, and TRBP2 and amino acid sequence alignment of the RRM and dsRBD from proteins from A. thaliana (At) and C. reinhardtii (Cr). Alignments were performed with ClustalX2. (A) Schematic domain structure of RNA-binding proteins involved in miRNA processing in C. reinhardtii (DUS16), A. thaliana (HYL1), and Homo sapiens (DGCR8 and TRBP2). Blue box, ssRNA recognition motif (RRM). Red box, dsRBD. (B) Alignment of RRMs. The conserved RNP1 and RNP2 sequences are indicated by black lines below the alignment (42, 43). (C) Alignment of dsRBDs. Amino acid residues conserved in animal polypeptides are indicated below the alignment (44, 45).