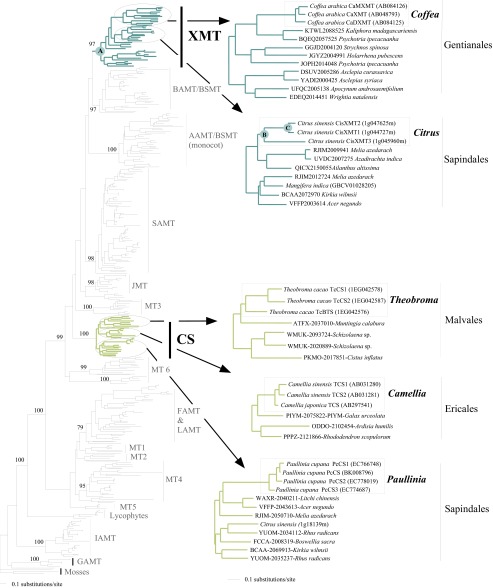
Fig. S1.
Phylogenetic relationships among 356 SABATH protein sequences. Sequences were extracted from 11 complete genomes of land plants in addition to selected CS and XMT transcriptome sequences from the oneKP database. Lineages with functionally characterized sequences are labeled by enzyme name, whereas those without known functions are arbitrarily numbered from MT1 to MT6. Bootstrap support values are shown for selected nodes that define major enzyme lineages. Enzymes from Camellia (CS) or Coffea (XMT) known to be involved in caffeine biosynthesis are shown in lime-green and turquoise, respectively. Sequences expressed in Theobroma and Paullinia fruits are clearly orthologous to CS sequences from Camellia. Sequences expressed in Citrus flowers are clearly orthologous to XMT sequences of Coffea. Arrows point to CS and XMT lineages to show recent duplication events within Theobroma, Paullinia, Camellia, Citrus, and Coffea. Nodes for which ancestral resurrected proteins were studied are labeled A–C. Accession numbers for the oneKP and GenBank databases are shown before and after relevant sequences, respectively.