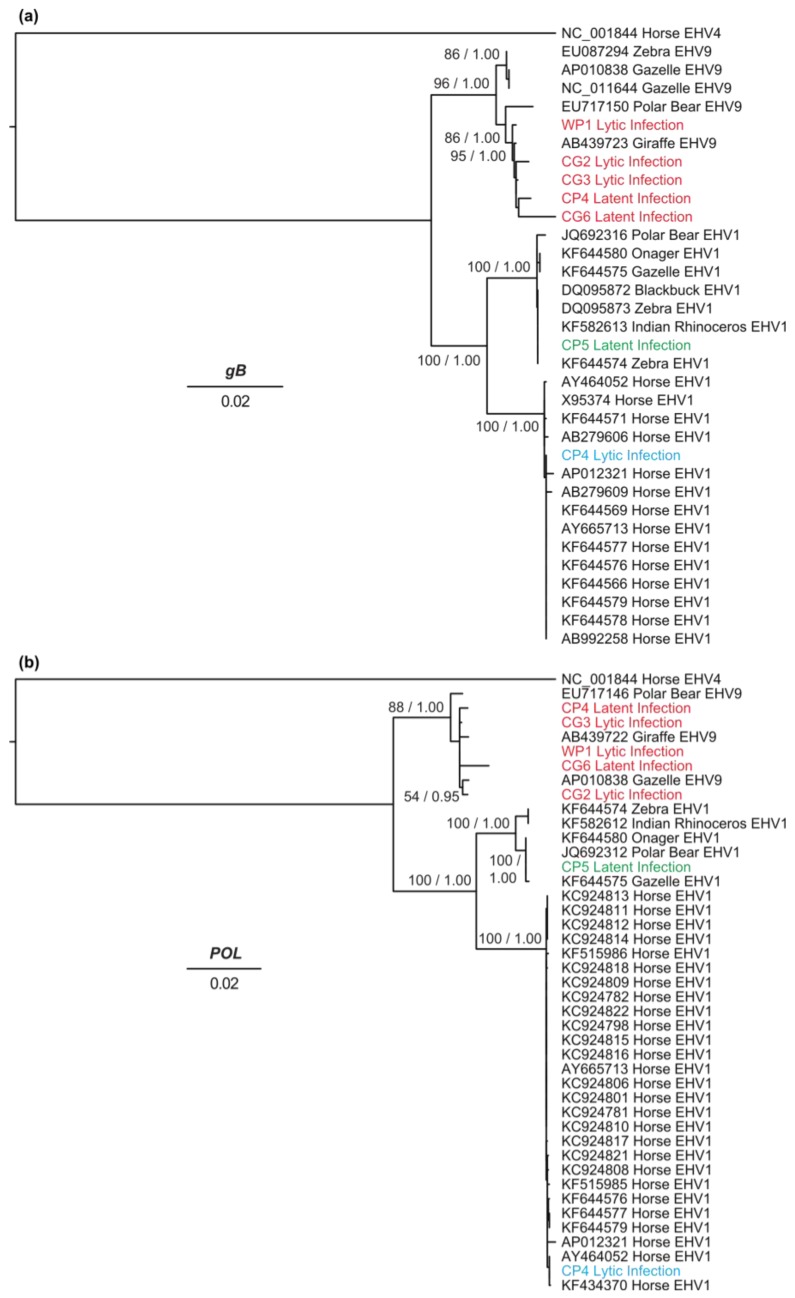
Figure 3.
Phylogenetic trees inferred using maximum likelihood from nucleotide sequences of (a) gB and (b) Pol genes for the six zebras WP1, CG2, CG3 (Equine herpesvirus 9 (EHV-9) lytic infection), CP4 (Equine herpesvirus 1 (EHV-1) lytic and EHV-9 latent infection), CP5, and CG6 (EHV-1 and EHV-9 latent infection, respectively) and other equine herpesviruses. Reference sequences are indicated by GenBank accession number, species from which the sequence was isolated, and viral strain. The novel EHV-9 sequences are in red, the novel EHV-1-horse like zebra sequence is in blue, and the novel zebra-EHV-1 sequence is in green. The trees are shown with branches lengths scaled to nucleotide substitutions per site. Selected nodes are labeled with maximum-likelihood bootstrap support values and posterior probabilities, separated by a slash “/”.