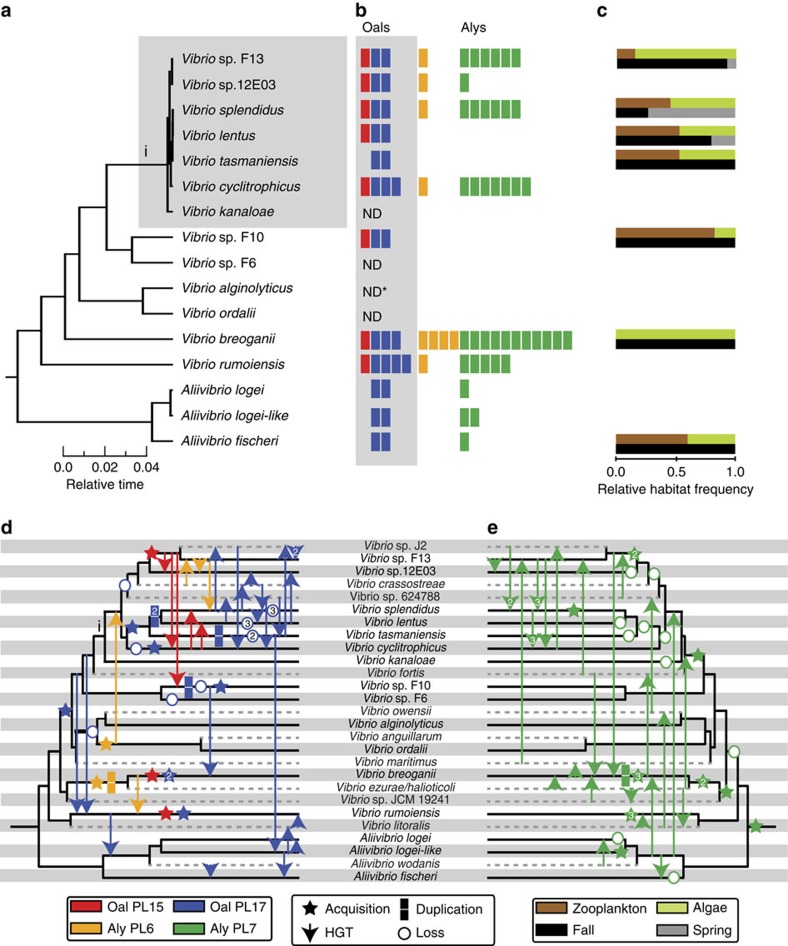
Figure 1. Evolutionary history and ecological occurrence of alginate lyases.
(a) Relative timed maximum-likelihood phylogeny of Vibrionaceae populations co-occurring in the same water samples. Species names are assigned if a previously described type strain falls within the population; otherwise, the designation Vibrio sp. is given. (b) Maximum copy number of alginate lyase families within members of a population identified by presence of a single enzymatic domain are represented by coloured rectangles. ND indicates that Alys and Olys were not detected. ND* indicates that while no Alys or Olys were detected in V. alginolyticus isolates from our collection, it is present in other V. alginolyticus isolates from geographically distant locations (Supplementary Fig. 1). (c) Normalized distribution of isolates obtained from algal detritus particles and zooplankton handpicked under a dissecting microscope and phylogenetically categorized by multilocus gene analysis for two seasonal samples. (d,e) Phylogenetic reconciliation (Methods) by comparison of pathway-specific gene trees (Supplementary Figs 6–9) and a timed ‘species' tree showing the history of each of four lyase gene families embedded in the reference species phylogeny: (d) Oal domains PL15 and PL17, and Aly domain PL6; (e) Aly domain PL7. Acquisition represents an independent entry of a subfamily into a clade within our collection. Solid and dashed lines on the phylogenetic tree indicate clades represented in our collection or obtained from Genbank, respectively. Numbers within symbols indicate multiple independent occurrences of the represented event. Within-population HGT and duplication are not depicted. Lowercase Roman numeral i indicates the crown group consisting of seven closely related populations.