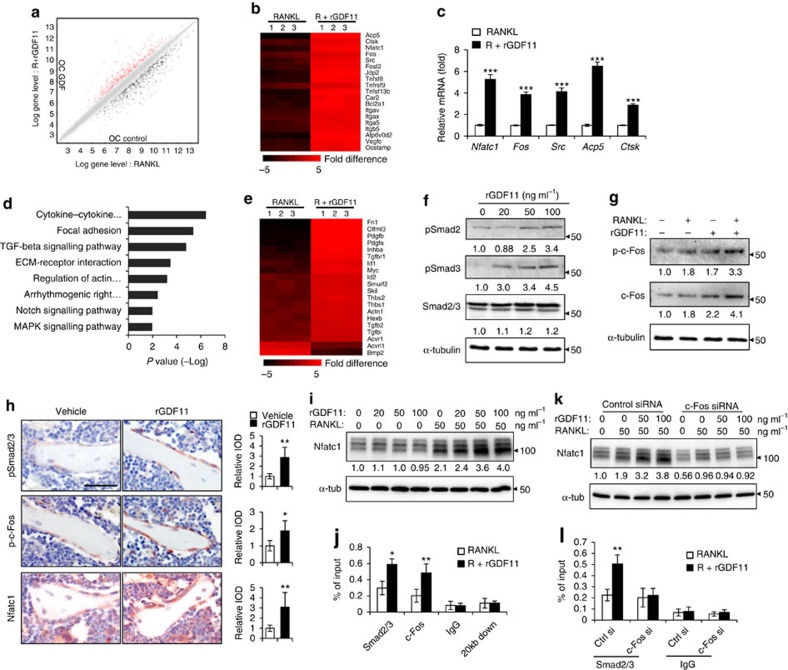
Figure 4. GDF11 activates Smad2/3-dependent TGF-β pathway.
(a) Gene expression levels of the BMMs stimulated without or with 100 ng ml−1 rGDF11 for 24 h (three biological replicates per group). 467 genes (red) were upregulated and 679 genes (black) were downregulated. 100 ng ml−1 M-CSF was present in all settings. (b) Heatmap of the osteoclastogenesis associated genes. (c) Quantitative RT-PCR confirmed the increased expression of osteoclast key marker genes. Results are shown as mean±s.d.; n=3. ***P<0.001 by t test. (d) KEGG pathway analysis indicated the altered function of TGF-β pathway. (e) Heatmap of the TGF-β pathway associated genes. (f) Western blot analysis indicated that rGDF11 stimulated the phosphorylation of Smad2/3 in BMMs in 30 min. (g) Western blot analysis demonstrated that rGDF11 amplified the RANKL-induced expression of c-Fos. BMMs were starved overnight and then treated for 4 h. (h) Representative images of immunohistochemical staining. rGDF11 injections increased the phosphorylation of Smad2/3 and c-Fos, as well as the expression of Nfatc1 in vivo. Femurs were collected ∼2 h after the last injection of rGDF11. Scale bar, 50 μm. (i) Western blot analysis indicated that rGDF11 stimulated the RANKL-induced expression of Nfatc1. BMMs were treated for 2 days. (j) ChIP assay revealed that rGDF11 induced the co-occupancy of Smad2/3 and c-Fos to the binding region of Nfatc1. Results are shown as mean±s.d.; n=3. *P<0.05 and **P<0.01 by t test. (k) Western blot analysis of Nfatc1. Depletion of c-Fos eliminated the rGDF11 induced expression of Nfatc1. (l) ChIP assay. Depletion of c-Fos abolished rGDF11 triggered binding of Smad2/3 to Nfatc1. Results are shown as mean±s.d.; n=3. **P<0.01 by t test.