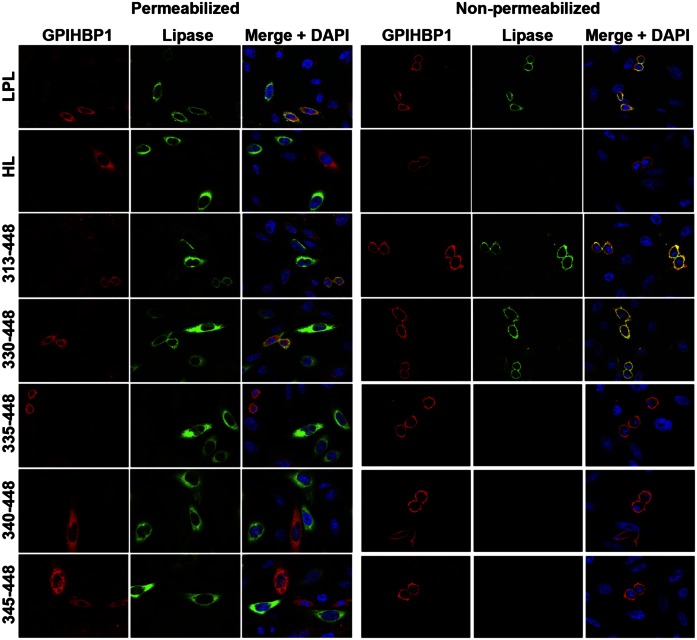
Fig. 7.
Immunofluorescence microscopy studies to assess the ability of LPL, HL, and LPL–HL chimeras to bind to GPIHBP1. CHO-K1 cells were electroporated with an S-protein–tagged human GPIHBP1 construct or a V5-tagged lipase expression vector (either LPL, HL, or HL–LPL chimeras containing LPL residues 313–448, 330–448, 335–448, 340–448, and 345–448) (see Fig. 1 for description of the constructs). The separately transfected cells were then mixed and plated on coverslips in 24-well plates. Twenty-four hours later, the cells were fixed in 3% paraformaldehyde, and blocked with 10% donkey serum in PBS/Mg/Ca. Some cells were permeabilized with 0.2% Triton X-100. Cells were then incubated for 1 h in blocking buffer with a goat polyclonal antibody against the S-protein tag (red) and a mouse mAb against the V5 tag (green), followed by a 30-min incubation with an Alexa 568–conjugated donkey anti-goat IgG (Invitrogen; 1:800) and an Alexa 488–conjugated donkey anti-mouse IgG (Invitrogen; 1:800). Cell nuclei were visualized with DAPI (blue). Cells expressing wild-type GPIHBP1 captured LPL secreted by neighboring LPL-expressing cells; hence, the GPIHBP1 and LPL signals colocalized on the merged image. HL–LPL (313–448) and HL–LPL (330–448) bound to GPIHBP1. HL and the remaining HL–LPL chimeras did not bind to GPIHBP1 (no colocalization of GPIHBP1 and the lipase on the merged image). HL–LPL chimeras containing LPL residues 370–448, 380–448, and 389–448 also failed to bind to GPIHBP1.