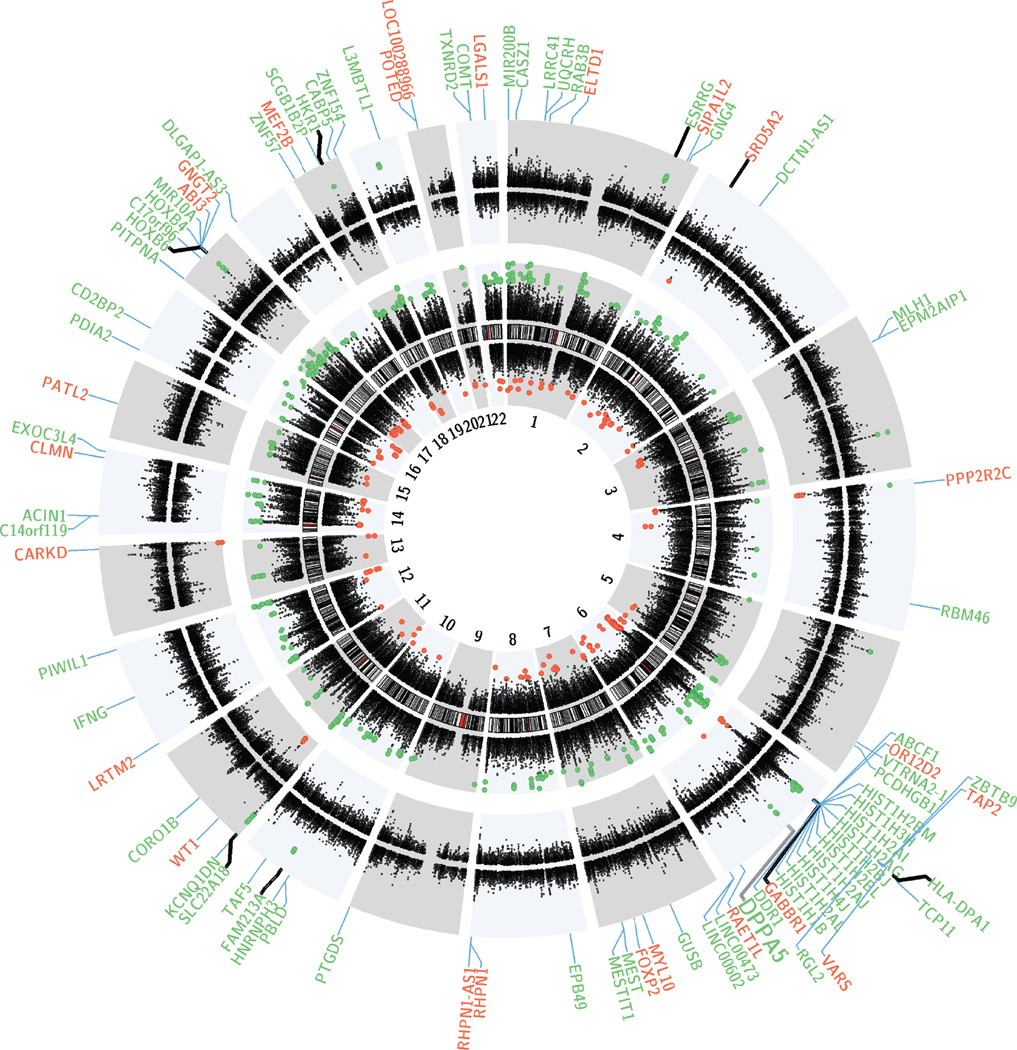
Figure 3. Genome-wide DNA methylation changes in previously malnourished G1 subjects and their G2 offspring.
Circos plot depicting the entire autosomal complement, green (red) genes with transcription start sites within ±2kb of significantly hyper-(hypo-)methylated DMRs. Genes with thin connector (Gene abbreviation to plot) line =G1; thick connector =G2. The black innermost ring (with vertical lines) represents autosome ideograms (annotated is the chromosomal number), with the pter-qter orientation in a clockwise direction. The dots on the inner side (negative) and outer side (positive) of the ideogram represent the Fisher’s method –log10 (P value) for each 1kb window analyzed in G1. Each dot marks the location of Illumina 450K probe along the genome. The next outermost black circle (with dots on the inner/negative and outer/positive side) represents the Fisher’s method –log10 (P value) for each 1kb window analyzed in G2. DPPA5 (green, larger font) was significantly hypermethylated both in G1 and G2 cohorts. Only genes where the DMR overlap its promoter i.e. ±2kb of TSS are shown.