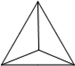
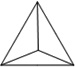
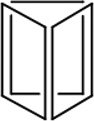
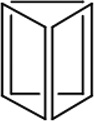
Table 1.
Physiochemical Properties and Stability of DNA Nanostructures.
 |
 |
 |
 |
 |
 |
 |
 |
 |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Length of edges |
~ 7 nm | ~ 7 nm | ~ 7 nm | ~ 7 nm | ~ 7 nm | ~ 7 nm | 90 × 60 nm | 120 nm (side width ~ 30 nm) |
16 × 16 × 30 nm |
80 × 20 nm |
| Further treatment after assembly |
Hexaethylene glycol (HEG) at the end of each component strand |
T4 ligase | Decorated with CpG sequences |
|||||||
| Incubation medium |
10% FBS |
50% FBS |
10% FBS | 10% FBS | 10% FBS | 10% FBS/PBS |
Cell lysates | Cell lysates | Cell lysates | FBS containing medium |
| Mean lifetime |
42 h | 24 h | 18 h | 62 h | 200 h | 3.5 h | 12 h | 12 h | 12 h | > 9 h |
| Control | dsDNA | dsDNA | ssDNA | HEG-ssDNA | ssDNA treated with T4 ligase |
dsDNA | ssDNA & dsDNA |
ssDNA & dsDNA |
ssDNA & dsDNA |
Template DNA |
| Control’s mean lifetime |
0.8 h | < 2 h | < 1 h | ~ 28 h | ~ 39 h | 0.7 h | < 1 h | < 1 h | < 1 h | ~ 6 h |
| Reference | 65 | 66 | 66 | 66 | 66 | 67 | 68 | 70 | 70 | 64 |
