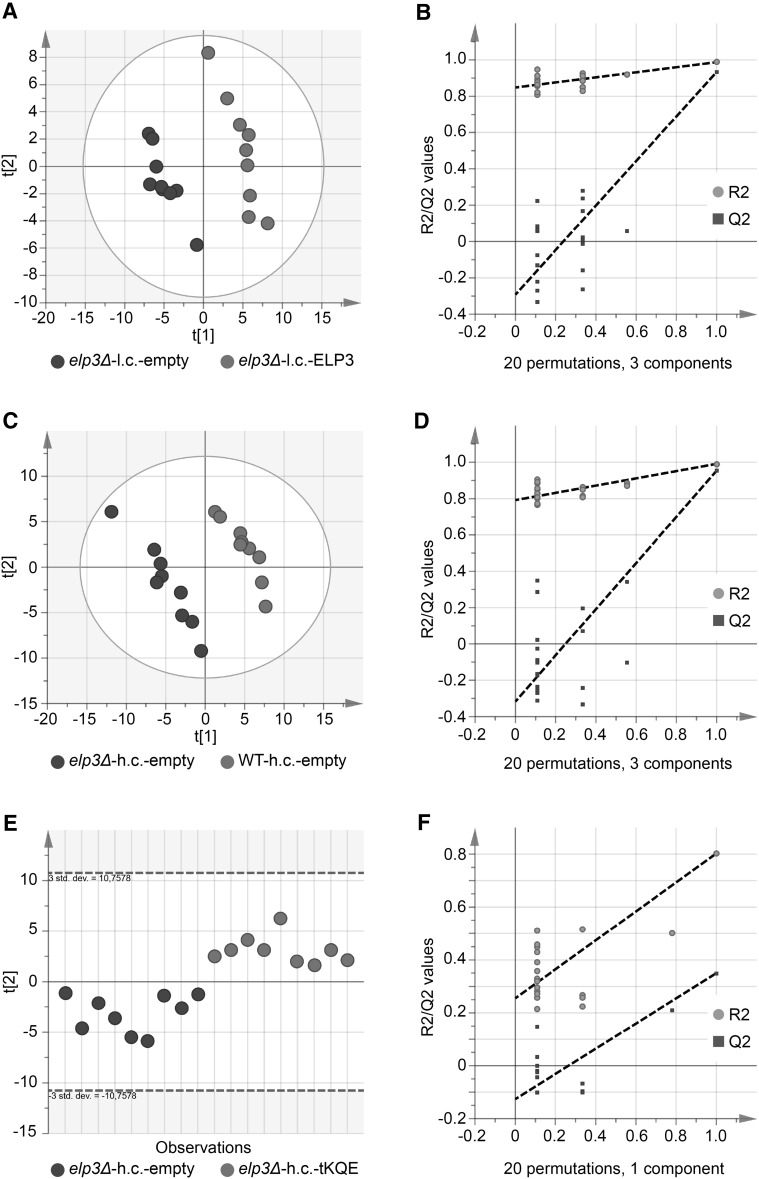
Fig. 3.
Score plots summarizing the PLS-DA modelling of strains grown at 30 °C. a PLS-DA score plot when modelling the elp3Δ strain with an empty low copy pRS315 vector (elp3Δ-l.c.-empty) against the elp3Δ strain containing the wild-type ELP3 gene on a pRS315 vector (elp3Δ-l.c.-ELP3). b Random permutation (20 randomizations) test-validation plot of PLS-DA model in (a). c PLS-DA score plot when modelling the elp3Δ strain with an empty high copy pRS425 vector (elp3Δ-h.c.-empty) against the wild-type strain with an empty pRS425 vector (WT-h.c.-empty). d Random permutation (20 randomizations) test-validation plot of PLS-DA model in (c). e PLS-DA score plot when modelling the elp3Δ strain with an empty high copy pRS425 vector (elp3Δ-h.c.-empty) against the elp3Δ strain containing a pRS425 vector carrying the tRNA genes tK(UUU), tQ(UUG) and tE(UUC) (elp3Δ-h.c.-tKQE). f Random permutation (20 randomizations) test-validation plot of PLS-DA model in (e). The PLS-DA model in e only has one valid component and is therefore portrayed by one vector. Each dot in a, c and e represents a technical replicate from three different biological replicates