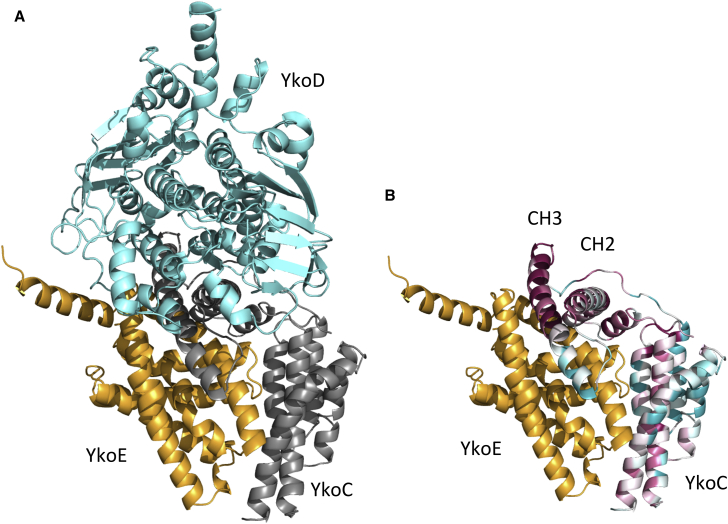
Figure 7.
Modeling of YkoEDC Complex
(A) Theoretical model of YkoEDC complex. YkoE is shown in orange, the homology model of YkoC in gray, and the homology model of YkoD in aquamarine.
(B) Theoretical model of YkoEC complex. YkoE is shown in orange, the homology model of YkoC colored according to conservation scores (cyan, variable; burgundy, conserved), the most conserved parts of YkoC are the helices CH2 and CH3, which are probably involved in interactions with YkoE and YkoD. An insertion and deletion in YkoC compared with EcfT could potentially result in some mechanistic differences between these T components. In YkoC, the loop connecting helix CH3 and transmembrane helix H5 is longer, and the hinge region between CH1 and CH2 is much shorter compared with the group II Ecf T component. The similarities in the pairwise alignments used to build the homology models are: YkoC and EcfT, 28%; YkoD N-terminal domain and EcfA2, 37%; YkoD C-terminal domain and EcfA1, 33% sequence identity.