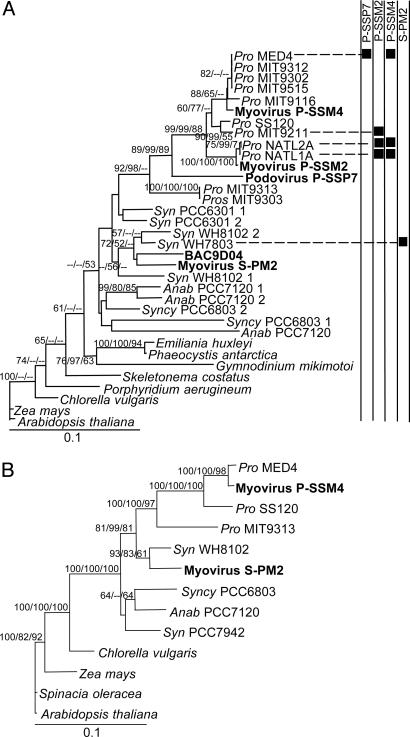
Fig. 2.
Distance trees of PSII core reaction center proteins. (A) D1 (psbA). (B) D2 (psbD). Phage sequences are shown in bold. The host strains that each phage infects are indicated by black squares. Trees were generated from 244 and 336 amino acids for D1 and D2, respectively (see Figs. 4 and 5). Bootstrap values for distance and maximum parsimony analyses and quartet puzzling values for maximum likelihood analysis >50% are shown at the nodes (distance/maximum likelihood/maximum parsimony). Trees were rooted with Arabidopsis thaliana proteins. Essentially, the same topology was obtained when nucleotide trees (third position excluded) were constructed, except for psbA from P-SSP7, which clustered with HL Prochlorococcus, albeit with low bootstrap support. Pro, Prochlorococcus; Syn, Synechococcus; Anab, Anabaena; Syncy, Synechocystis.